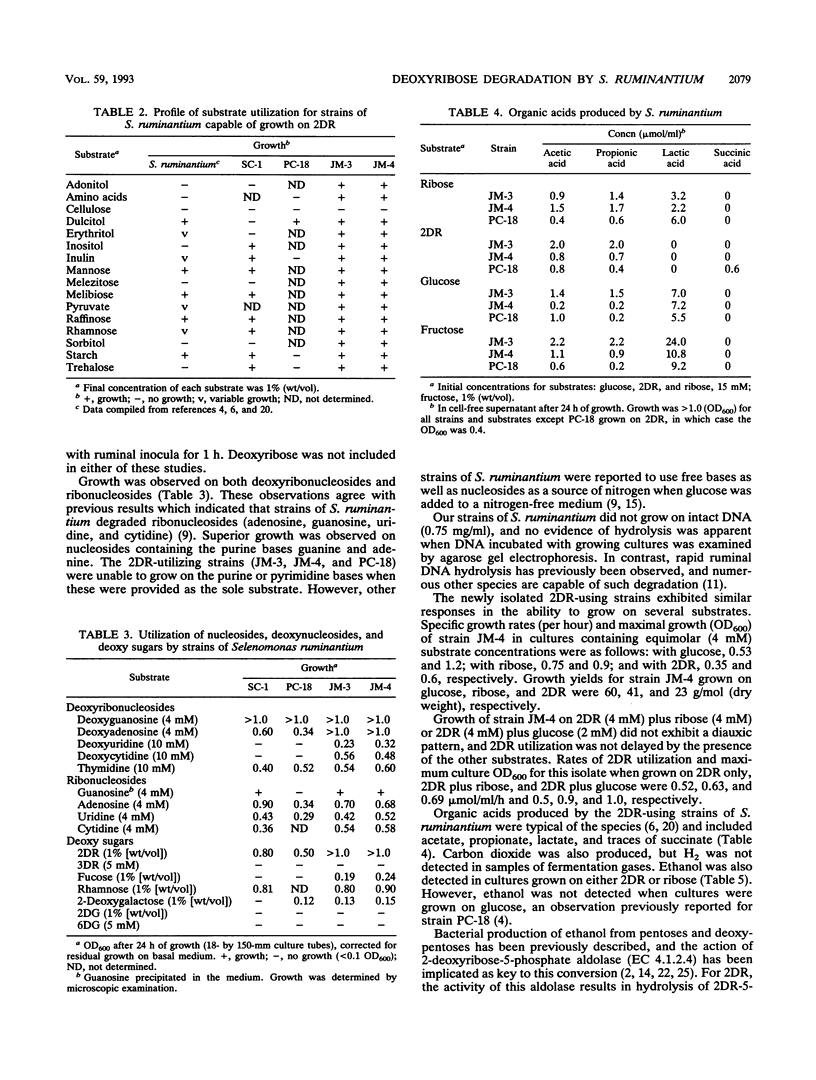
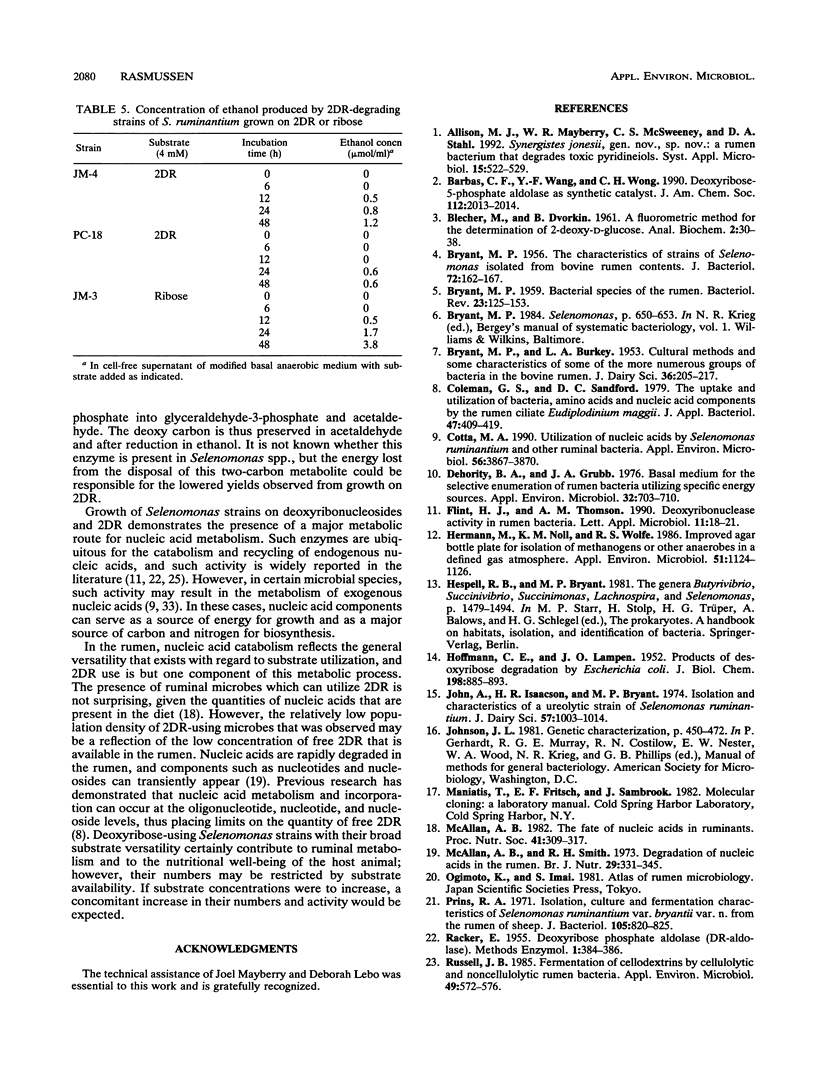
Abstract
Microbes from ruminal contents of cattle were selectively enriched by using 2-deoxyribose (2DR) as a substrate for growth. Bacterial isolates growing on 2DR were gram-negative, curved, motile rods. The isolates grew on a broad range of substrates, including deoxyribose, glucose, ribose, mannitol, and lactate as well as ribonucleosides and deoxyribonucleosides. The strains also grew on rhamnose (6-deoxymannose) but not DNA. Organic acids produced from growth on hexoses and pentoses included acetate, propionate, lactate, and succinate. The isolates were identified as Selenomonas ruminantium subsp. lactilytica on the basis of morphology, substrate specificity, and other biochemical characteristics. Several characterized species of ruminal bacteria were also screened for growth on 2DR, with only one strain (S. ruminantium PC-18) found able to grow on 2DR. Ethanol was produced by 2DR when strains were grown on ribose or 2DR.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRYANT M. P. Bacterial species of the rumen. Bacteriol Rev. 1959 Sep;23(3):125–153. doi: 10.1128/br.23.3.125-153.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRYANT M. P. The characteristics of strains of Selenomonas isolated from bovine rumen contents. J Bacteriol. 1956 Aug;72(2):162–167. doi: 10.1128/jb.72.2.162-167.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta M. A. Utilization of nucleic acids by Selenomonas ruminantium and other ruminal bacteria. Appl Environ Microbiol. 1990 Dec;56(12):3867–3870. doi: 10.1128/aem.56.12.3867-3870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehority B. A., Grubb J. A. Basal medium for the selective enumeration of rumen bacteria utilizing specific energy sources. Appl Environ Microbiol. 1976 Nov;32(5):703–710. doi: 10.1128/aem.32.5.703-710.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint H. J., Thomson A. M. Deoxyribonuclease activity in rumen bacteria. Lett Appl Microbiol. 1990 Jul;11(1):18–21. doi: 10.1111/j.1472-765x.1990.tb00126.x. [DOI] [PubMed] [Google Scholar]
- HOFFMANN C. E., LAMPEN J. O. Products of desoxyribose degradation by Escherichia coli. J Biol Chem. 1952 Oct;198(2):885–893. [PubMed] [Google Scholar]
- Hermann M., Noll K. M., Wolfe R. S. Improved agar bottle plate for isolation of methanogens or other anaerobes in a defined gas atmosphere. Appl Environ Microbiol. 1986 May;51(5):1124–1126. doi: 10.1128/aem.51.5.1124-1126.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John A., Isaacson H. R., Bryant M. P. Isolation and characteristics of a ureolytic strain of Selenomonas ruminatium. J Dairy Sci. 1974 Sep;57(9):1003–1014. doi: 10.3168/jds.s0022-0302(74)85001-0. [DOI] [PubMed] [Google Scholar]
- McAllan A. B., Smith R. H. Degradation of nucleic acids in the rumen. Br J Nutr. 1973 Mar;29(2):331–345. doi: 10.1079/bjn19730107. [DOI] [PubMed] [Google Scholar]
- McAllan A. B. The fate of nucleic acids in ruminants. Proc Nutr Soc. 1982 Sep;41(3):309–317. doi: 10.1079/pns19820046. [DOI] [PubMed] [Google Scholar]
- Prins R. A. Isolation, culture, and fermentation characteristics of Selenomonas ruminantium var. bryantivar. n. from the rumen of sheep. J Bacteriol. 1971 Mar;105(3):820–825. doi: 10.1128/jb.105.3.820-825.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Baldwin R. L. Substrate preferences in rumen bacteria: evidence of catabolite regulatory mechanisms. Appl Environ Microbiol. 1978 Aug;36(2):319–329. doi: 10.1128/aem.36.2.319-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B. Fermentation of cellodextrins by cellulolytic and noncellulolytic rumen bacteria. Appl Environ Microbiol. 1985 Mar;49(3):572–576. doi: 10.1128/aem.49.3.572-576.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgarrella F., Del Corso A., Tozzi M. G., Camici M. Deoxyribose 5-phosphate aldolase of Bacillus cereus: purification and properties. Biochim Biophys Acta. 1992 Jan 9;1118(2):130–133. doi: 10.1016/0167-4838(92)90139-5. [DOI] [PubMed] [Google Scholar]
- Stanton T. B., Canale-Parola E. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch Microbiol. 1980 Sep;127(2):145–156. doi: 10.1007/BF00428018. [DOI] [PubMed] [Google Scholar]
- Strobel H. J., Russell J. B. Succinate transport by a ruminal selenomonad and its regulation by carbohydrate availability and osmotic strength. Appl Environ Microbiol. 1991 Jan;57(1):248–254. doi: 10.1128/aem.57.1.248-254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R. M. Maintenance of Laboratory strains of obligately anaerobic rumen bacteria. Appl Environ Microbiol. 1982 Aug;44(2):499–501. doi: 10.1128/aem.44.2.499-501.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner K. W., Roberton A. M. Xylose, arabinose, and rhamnose fermentation by Bacteroides ruminicola. Appl Environ Microbiol. 1979 Jul;38(1):7–12. doi: 10.1128/aem.38.1.7-12.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varel V. H. Reisolation and characterization of Clostridium longisporum, a ruminal sporeforming cellulolytic anaerobe. Arch Microbiol. 1989;152(3):209–214. doi: 10.1007/BF00409652. [DOI] [PubMed] [Google Scholar]


