Abstract
Current ambient UV-B levels can significantly depress productivity in aquatic habitats, largely because UV-B inhibits several steps of photosynthesis, including the photooxidation of water catalyzed by photosystem II. We show that upon UV-B exposure the cyanobacterium Synechococcus sp. PCC 7942 rapidly changes the expression of a family of three psbA genes encoding photosystem II D1 proteins. In wild-type cells the psbAI gene is expressed constitutively, but strong accumulations of psbAII and psbAIII transcripts are induced within 15 min of moderate UV-B exposure (0.4 W/m2). This transcriptional response causes an exchange of two distinct photosystem II D1 proteins. D1:1 is encoded by psbAI, but on UV-B exposure, it is largely replaced by the alternate D1:2 form, encoded by both psbAII and psbAIII. The total content of D1 and other photosystem II reaction center protein, D2, remained unchanged throughout the UV exposure, as did the content and composition of the phycobilisome. Wild-type cells suffered only slight transient inhibition of photosystem II function under UV-B exposure. In marked contrast, under the same UV-B treatment, a mutant strain expressing only psbAI suffered severe (40%) and sustained inhibition of photosystem II function. Another mutant strain with constitutive expression of psbAII and psbAIII was almost completely resistant to the UV-B treatment, showing no inhibition of photosystem II function and only a slight drop in electron transport. In Synechococcus the rapid exchange of alternate D1 forms, therefore, accounts for much of the cellular resistance to UV-B inhibition of photosystem II activity and photosynthetic electron transport. This molecular plasticity may be an important element in community-level responses to UV-B, where susceptibility to UV-B inhibition of photosynthesis changes diurnally.
Keywords: photosynthesis, phycobiliprotein, psbA
In oxygenic photobionts, photosystem II (PSII) is an integral membrane complex that catalyzes the photooxidation of water, with concomitant release of oxygen. Electrons extracted from water are passed to plastoquinone and enter the photosynthetic electron transport chain. The core of the PSII complex is composed of a dimer of two related proteins, D1 and D2, that bind the pigments and cofactors involved in this electron transfer from water to plastoquinone. During active photosynthesis the D1 protein, and to a lesser extent D2, turn over rapidly and are replaced by newly synthesized polypeptides in a PSII repair cycle. Under environmental stress, the repair cycle can be impaired, such that degradation and loss of D1 protein exceeds the rate of replacement (1). This net loss of functional D1 leads to a drop in PSII function and can contribute to photoinhibition, a light-dependent drop in the quantum yield of photosynthesis. Photoinhibition usually occurs when excitation capture exceeds the rate of electron removal from the PSII complex, as can occur when the light intensity exceeds the acclimated irradiance or when the temperature drops below the acclimated level.
UV-B absorption also leads to photoinhibition of both isolated PSII preparations and intact cells, via damage to the D1 protein or associated cofactors, which triggers degradation of the D1 protein (2–7). UV-B also inhibits other photosynthetic functions, including expression and activity of ribulose-1,5-bisphosphate carboxylase/oxygenase (8–12), the key enzyme of carbon dioxide fixation. In cyanobacteria, UV-B can trigger dissociation and bleaching of the phycobilisome light-harvesting antennae (12–16), inhibit nitrogenase activity (17–19), and interfere with cell differentiation (20), although the ecological significance of some severe UV treatments has been questioned (21).
Known chloroplasts contain only one active gene, psbA, encoding the D1 protein. In contrast, all cyanobacteria studied to date contain multigene psbA families whose expression is regulated in response to environmental conditions (22–25). The cyanobacterium Synechococcus sp. PCC 7942 contains a family of three psbA genes encoding two distinct forms of the PSII D1 protein (24). During acclimated growth, transcripts from psbAI predominate and are translated to produce the D1 form 1 protein (D1:1). When the cells suffer excess excitation psbAI transcripts are rapidly replaced by transcripts from psbAII and psbAIII, encoding D1:2 (24, 26). This leads to rapid exchange of D1:1 for D1:2 within PSII (27, 28), with D1:2 remaining the predominant form until cells acclimate to their new growth regime, at which time D1:2 is again replaced by D1:1 (29). The resulting transient exchange of D1 forms appears to be triggered by a fractional closure of PSII centers (30). The exchange between D1:1 and D1:2 is an essential element of the Synechococcus response to increased light or decreased temperature, because if the exchange is blocked, the cells suffer severe and sometimes irreversible photoinhibition and are impaired in their ability to eventually acclimate to the new conditions (27).
Because the psbA/D1 exchange system appears central to cyanobacterial resistance to excitation-induced photoinhibition, we decided to test whether the system is also important for cellular resistance to moderate UV-B. Over their long evolution, cyanobacteria have survived many different UV-B regimes (31), and currently natural UV-B levels are increasing in aquatic habitats, with unknown long-term implications for productivity and community structure (21, 32–36). We show that moderate levels of UV-B induce rapid and extensive expression of the psbAII and psbAIII genes in Synechococcus sp. PCC 7942, leading to complete replacement of the D1:1 protein with D1:2. Furthermore, we used gene-inactivation mutants lacking either D1:1 or D1:2 to demonstrate that the exchange between D1:1 and D1:2 is essential for limiting the extent of UV-induced photoinhibition in Synechococcus sp. PCC 7942.
MATERIALS AND METHODS
Culture Growth Conditions.
Synechococcus sp. PCC 7942 batch cultures of 300 ml were grown in flat rectangular flasks (culture depth of about 1 cm) in BG-11 inorganic medium (37), supplementally buffered with 10 mM 3-(N-morpholino)propanesulfonic acid (pH 7.5), and bubbled with 5% CO2 in air (about 1 ml/s) at 37°C under continuous even incandescent illumination of 50 μmol of photons per m2 per s. Chlorophyll (Chl) and phycocyanin content were determined by using whole cell spectra (38). Cultures were inoculated with liquid preculture to a concentration of about 0.5 μg of Chl per ml and used for UV-B treatments when they reached about 2 μg of Chl per ml, in the exponential growth phase.
UV-B Treatments.
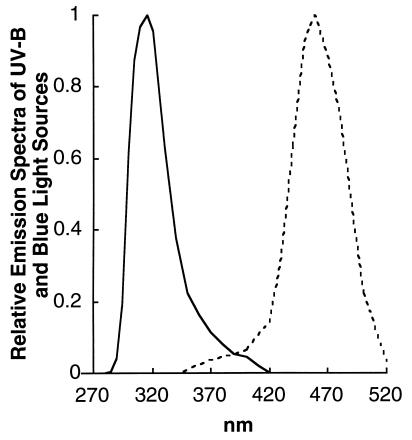
Fluorescent UV-B lamps (20W/12, Philips Lighting, Stockholm) were used to irradiate cultures in polystyrene flasks (Nunclon, Nalge). The UV-B irradiation spectrum (Fig. 1) was calculated from the lamp emission and the transmission of the plastic flask. The flasks had zero transmission below 280 nm, and so the UV-C range was excluded. UV-B intensity was measured with a model SKU430 UV-B (280–315 nm) light sensor (Skye Instruments, Llandrindod Wells, Powys, Wales, U.K.). The surface of the cell culture was exposed to UV-B at 0.2, 0.4, or 0.8 W/m2 (0.5, 1, and 2 μmol of UV-B per m2 per s, respectively) with continued incandescent growth light of 50 μmol of photons per m2 per s. For blue light treatments, a fluorescent tube wrapped in a blue filter (Fig. 1) was used to provide 5 μmol of photons per m2 per s (about 1.3 W/m2) to supplement the 50 μmol of photons per m2 per s from the incandescent growth light.
Figure 1.
Relative spectral irradiance profiles of UV-B and blue light sources. UV-B, solid curve; blue light, dashed curve. The culture flasks blocked wavelengths less than 280 nm and thus the UV-C range was excluded. In all treatments, an incandescent growth light of 50 μmol of photons per m2 per s was maintained. UV-B treatments added UV-B at culture surface intensities of 0.2, 0.4, or 0.8 W/m2 (about 0.5, 1, and 2 μmol of UV-B per m2 per s, respectively). Blue light treatments added culture surface intensities of 5 μmol of photons per m2 per s (about 1.3 W/m2).
Detection of psbA Messages.
Total RNA was isolated from Synechococcus sp. PCC 7942 (30) and 3 μg was denatured by glyoxylation (39), electrophoretically fractionated in 1% agarose gels in 10 mM sodium phosphate (pH 7.0), and then transferred to Hybond-N membrane (Amersham). Transcripts of all three psbA genes were then detected by using DNA probes specific for the unique 5′ untranslated regions of each gene (100 bp) (30). The amount of hybridization to each psbA transcript was determined directly from the filter by using a PhosphoImager (Bio-Rad) and quantified relative to the amount of 16S RNA (40).
Protein Detection by Immunoblotting.
Total cellular proteins were extracted (27) and samples containing 0.75 μg of Chl were separated on linear lithium dodecyl sulfate/15% polyacrylamide gels. Proteins were then electrophoretically transferred to poly(vinylidene difluoride) (0.2-μm pore size, Immobilon-P, Millipore), and immunoblot analysis was performed (41), with detection by using the ECL chemiluminescent kit (Amersham). The polyclonal antibodies against D1:1 and D1:2 are completely form-specific and show no detectable cross-reactivity (27). The antibodies against D1, D2, and total phycobilisome polypeptides are described elsewhere (29, 42).
Photosynthetic Measurements.
Chl a fluorescence yield and oxygen evolution were measured simultaneously with an oxygen electrode (Hansatech Instruments, Pentney King’s Lynn, England) and a pulse-amplitude-modulated Chl fluorometer (Walz, Effeltrich, Germany) (29). The relative efficiency of excitation energy capture by PSII reaction centers was estimated as (FM − FO)/FM (43).
RESULTS AND DISCUSSION
UV-B Strongly Regulates Expression of psbA Genes and Leads to Exchange of Alternate PSII D1 Proteins.
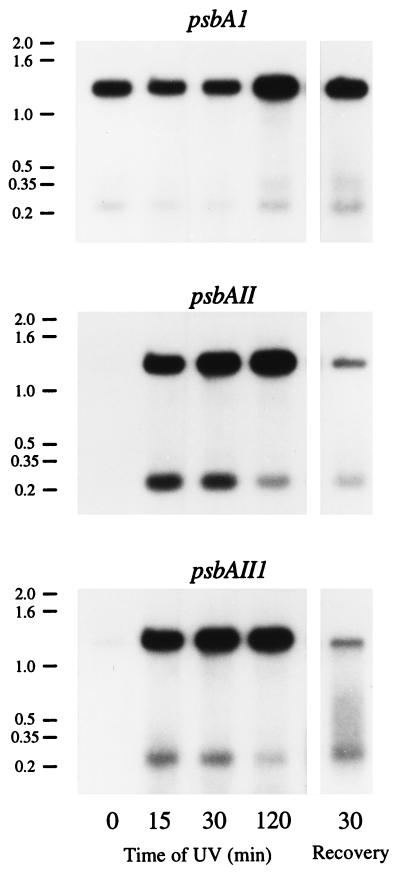
Figs. 2 and 3A show a representative hybridization of psbA messages with gene-specific probes and their average quantification in two replicates, respectively. For the quantification, we assumed the composition of psbA mRNA under control growth conditions was 94% psbAI, 1.4% psbAII, and 4.6% psbAIII (24, 30). In wild-type Synechococcus cells acclimated to moderate light, the psbAI gene encoding the PSII protein D1:1 is expressed constitutively (refs. 24 and 44 and Fig. 2). There is little expression of the psbAII and psbAIII genes (Figs. 2 and 3A), encoding the alternate D1:2, and thus the PSII centers contain almost exclusively D1:1 (refs. 27, 29, 30, 40, and 45 and Fig. 4). We supplemented the white growth light (50 μmol per m2 per s) with a moderate UV-B treatment (Fig. 1, 0.4 W/m2 = 1 μmol of UV-B incident on surface per m2 per s), chosen as an ecologically reasonable level (3, 4, 32, 35, 46).
Figure 2.
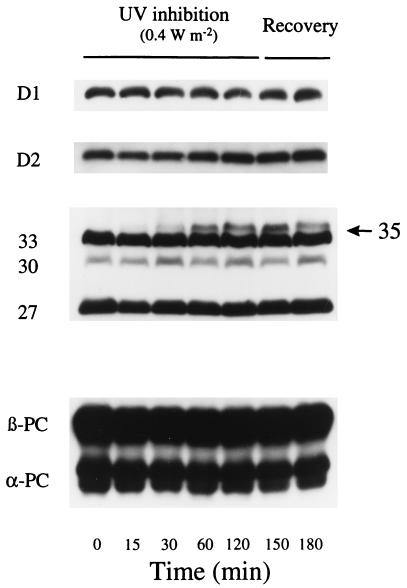
Differential expression of psbA transcripts after moderate UV-B irradiation supplemented to the standard growth light. Synechococcus sp. PCC 7942 wild-type cells grown at 50 μmol of photons per m2 per s were exposed to supplementary UV-B at 0.4 W/m2 for 2 h and then allowed to recover without UV-B for 30 min. Cell samples were taken at the indicated times for RNA isolations. The levels of psbAI, psbAII, and psbAIII mRNA were detected by hybridization with 100-bp DNA probes specific for the unique 5′ untranslated region of each psbA transcript. Representative autoradiograms from one of two replicates are shown. Molecular size markers in kb are indicated on the left.
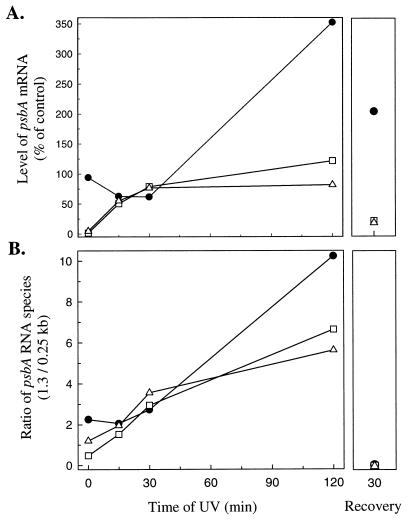
Figure 3.
Quantification of psbA transcripts under control, UV-B, and postexposure recovery treatments. (A) Levels of full-length 1.3-kb transcripts. psbAI, •; psbAII, □; psbAIII, ▵. Values plotted are the average of two independent replicates, expressed relative to the content at time zero. (B) Ratio of psbA mRNA species (1.3/0.25 kb). psbAI, •; psbAII, □; psbAIII, ▵. Values plotted are the average of two replicates.
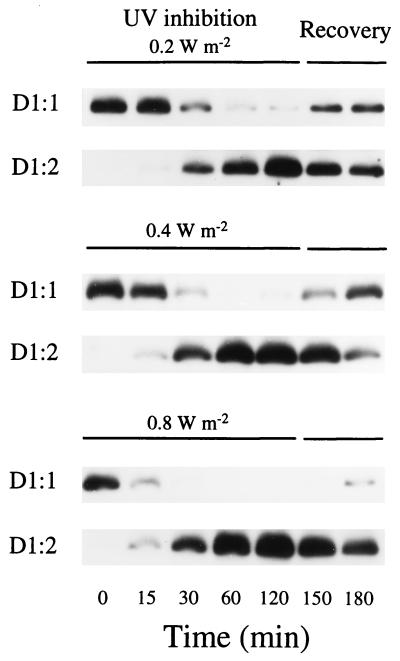
Figure 4.
Rapid exchange between D1:1 and D1:2 with increasing levels of supplemented UV-B irradiation and subsequent recovery. Synechococcus sp. PCC 7942 wild-type cells grown at 37°C with white light at 50 μmol of photons per m2 per s and bubbled with 5% CO2 were supplemented with UV-B at 0.2, 0.4, or 0.8 W/m2 for 2 h and then allowed to recover for 1 h without UV-B. Protein extracts were taken at the indicated times, and the composition of D1 protein was determined by immunoblotting with polyclonal antibodies specific for either D1:1 or D1:2. The figure shows results representative of three replicates.
Within 15 min of UV-B exposure the wild-type Synechococcus strongly induced accumulation of psbAII and psbAIII transcripts (Figs. 2 and 3A). Because the control cells contained almost no transcript or transcript fragments from psbAII and very little from psbAIII, this accumulation likely results largely from transcriptional induction, rather than changes in mRNA stability (Fig. 2). The DNA probes used recognize the unique 5′ untranslated regions of each gene and detect both the full-length transcripts of about 1.3 kb and also a smaller 0.25-kb cleavage product derived from the full-length transcript (24, 30). The 0.25-kb fragment for psbAII and psbAIII was relatively abundant during the first 15 min of UV-B exposure but then steadily declined, despite continued high levels of the mature transcripts (Fig. 2). The decline in the level of the 0.25-kb fragment from all three psbA transcripts results in a large increase in the ratio of 1.3/0.25-kb psbA RNA species (Fig. 3B). A similar pattern also occurs during chilling of Synechococcus (30) and may indicate progressive stabilization of the transcripts, which are relatively unstable when first transcribed (28, 30, 40).
The accumulation of psbAII and psbAIII transcripts leads to a gradual replacement of D1:1 with D1:2 in the PSII centers (Fig. 4). The exchange of the alternate D1 forms became faster and more complete as the UV-B intensity increased over a range from 0.2 to 0.8 W/m2 (Fig. 4). High levels of psbAI transcripts remained throughout the treatment (Fig. 2), indicating that posttranscriptional regulation favored D1:2 accumulation during UV-B exposure. The total cellular content of D1 protein (D1:1 + D1:2; Fig. 5) remained essentially constant, as did the content of D2, the other main PSII reaction center core protein (Fig. 5). During a recovery period, with cells incubated under control growth conditions of 50 μmol of photons per m2 per s, psbAII and psbAIII transcript levels rapidly declined, while psbAI transcripts remained (Fig. 2). This led to a recovery of D1:1 content and some drop in D1:2 content within 60 min of recovery (Fig. 4). Under prolonged UV exposure, the cells almost completely revert to the D1:1 protein, as other resistance mechanisms are induced (J. Porankiewicz and A. K. Clarke, personal communication).
Figure 5.
D1, D2, and phycobilisome protein content during moderate UV-B exposure and recovery. Wild-type cells were supplemented with UV-B at 0.4 W/m2 for 2 h and then allowed to recover for 1 h without UV-B as described in Fig. 3. Total D1 (D1:1 and D1:2), D2, and phycobilisome were detected by using specific polyclonal antibodies. The figure shows results representative of three replicates.
UV-B and Light Excitation Are Distinct Regulators of the psbA/D1 System.
Exposing Synechococcus cells to increased light or decreased temperature also causes exchange of the D1 forms (27–30, 45, 47), but this excitation stress response is distinct from the UV-B effects. Under excess excitation, psbAI expression is repressed, but transcription resumes as the cells acclimate to the new conditions of higher light or lower temperature (29, 30). The transient loss of psbAI message is compensated by a strong induction of psbAII and psbAIII expression, apparently triggered by PSII reduction or closure, when excitation capture exceeds the removal of electrons from the center (29, 30). In contrast, under UV-B irradiation psbAI transcripts are maintained and indeed increase above control levels (Fig. 3A) but are supplemented by transcription from psbAII and psbAIII. Under UV-B, total D1 content remains constant (Fig. 5), but the cells shift from D1:1 to D1:2 (Fig. 4). Therefore, posttranscriptional regulation appears to favor D1:2 accumulation, even though psbAI transcripts encoding D1:1 remain near or above the levels of psbAII and psbAIII transcripts (compare Figs. 2–4). Furthermore, during the recovery from UV-B, psbAII and psbAIII transcripts significantly decrease 30 min after cessation of UV-B (Figs. 2 and 3), and yet cells only slowly revert to the D1:1 protein despite a high level of psbAI transcript (Fig. 4). In contrast, during recovery from brief periods of excitation stress, cells revert to D1:1 almost immediately (29, 30). The supplemental expression of psbAII and psbAIII under UV-B is reminiscent of the light regulation of psbDII, a gene encoding an identical extra copy of the D2 protein of PSII, which is induced under excess light to supplement the constitutive expression of psbDI (48).
In nature, cells are only exposed to significant UV-B when light levels are also high. Therefore, the two factors, although mechanistically distinct, exert a combined evolutionary pressure to withstand both UV-B and light stress simultaneously. In this case, the psbA/D1 system helps cells cope with both stresses, but UV-B and light appear to trigger partly independent induction mechanisms. This is particularly true for the regulation of psbAI, which is transiently repressed by high light but not by UV-B.
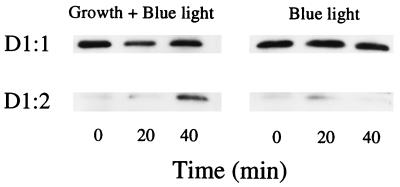
Blue light exposure induces some expression of psbAII and psbAIII through a regulatory system independent of PSII and electron transport (49). We found that blue light caused a small accumulation of D1:2 (Fig. 6), confirming that the blue light transcriptional response can cause a shift in protein composition. This weak blue light effect could reflect the extreme upper end of the UV response. Thus, the UV-B and blue light data suggest that UV-B is detected by a specific receptor, because UV-B and PSII overexcitation induce different patterns of gene and protein expression. Our results do not, however, rule out the possibility that the UV-B receptor is an element of photosynthetic electron transport. Interestingly, plastoquinone molecules in PSII have been postulated as receptors for UV-B-induced D1 degradation (2, 3) or PSII inhibition (50). These molecules have absorption peaks in the UV-B and blue regions, which gives raise to the hypothesis that plastoquinones are the receptors for the UV-B and blue light regulation of psbA expression, although there is evidence against quinones as the primary targets for UV-B inhibition of PSII and D1 degradation (50–52).
Figure 6.
D1:1 and D1:2 content in cells exposed to 50 μmol of white light per m2 per s (control growth conditions) supplemented with 6 μmol of blue light per m2 per s. The figure shows results representative of three replicates.
Moderate UV-B Has Little Effect on Phycobilisome Content.
The phycocyanin/Chl absorbance ratio dropped only slightly in the wild-type strain during the 2-h UV-B treatments, and the total phycobiliprotein content did not change (Fig. 5). High UV-B levels (e.g., 2.6 W/m2; ref. 12) can drive dissociation and photodestruction of isolated phycobilisomes (12, 13) and in intact cyanobacteria (14–16), but our data suggest these effects are minor in Synechococcus cells under moderate UV-B intensities. Interestingly, under UV-B exposure a new polypeptide of approximately 35 kDa was detected on the immunoblot with a polyclonal antibody raised against phycobilisomes from Synechococcus (Fig. 5). The antibody recognizes all the polypeptides from pure Synechococcus phycobilisomes, including the linker polypeptides, but does not cross-react with any other proteins from the cells (42). The new polypeptide, therefore, apparently shares antigenic determinants with normal constitutive phycobilisome polypeptides. The size suggests that it is a phycobilisome linker polypeptide, but the functional significance of this 35-kDa polypeptide is as yet unknown.
D1 Exchange Is Necessary for Resistance to UV-B Inhibition of Photosynthesis.
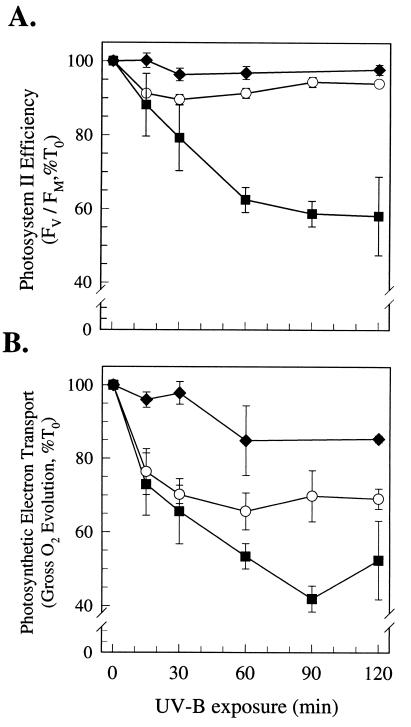
We next studied the biological significance of the exchange of D1:1 and D1:2 induced by UV-B. In wild-type cells, PSII function measured as the fluorescence ratio FV/FM suffered some initial inhibition (10%) upon exposure to UV-B (0.4 W/m2; Fig. 7A). This inhibition occurred over the same period when D1:1 protein was lost (Fig. 4). PSII function partially recovered and stabilized (Fig. 7A) as D1:2 accumulated (Fig. 4). Electron transport (measured as gross oxygen evolution) also suffered inhibition (Fig. 7B) but stabilized at 70% of the control level.
Figure 7.
Photosynthetic activity under moderate UV-B exposure. Wild-type cells (○), initially containing D1:1 but switching to D1:2 within 30 min, R2S2C3 mutant cells containing only D1:1 protein (▪), and R2K1 mutant cells containing only D1:2 protein (♦). (A) PSII function measured by using the Chl fluorescence parameter FV/FM. (B) Photosynthetic electron transport measured as gross oxygen evolution. Values are expressed as a percentage of the 0-h control (100%) and represent the mean ± SEM (n = 3).
In marked contrast, under the same UV-B treatment a mutant strain (R2S2C3, ref. 24) that expresses only the psbAI gene suffered severe and sustained inhibition. PSII efficiency dropped to 58% of control and electron transport fell to only 42% of control (Fig. 7). In this strain, the normal replacement of D1:1 with D1:2 is blocked because the psbAII and psbAIII genes are inactivated (24).
In the mutant strain R2K1, the psbAI gene is inactivated and the cells constitutively express the intact psbAII and psbAIII genes, and thus the cells contain only D1:2 (24). The R2K1 strain was highly resistant to the UV-B treatment (Fig. 7), showing no significant inhibition of PSII function and only a modest drop in electron transport to 85% of control after 120 min of UV-B exposure. Under control conditions, the R2K1 and R2S2C3 strains have similar levels of total D1 protein (27, 53).
Thus, the wild-type strain suffers progressive UV-B inhibition of PSII activity until D1:2 accumulates. If accumulation of D1:2 is blocked, as in the mutant strain R2S2C3, severe inhibition ensues. In contrast, constitutive expression of D1:2 confers strong resistance to UV-B inhibition.
The D1:1 and D1:2 proteins differ at 25 of 360 amino acid positions (24), including the change of amino acid Gln130 in D1:1 to Glu130 in D1:2, a position that interacts with the pheophytin redox cofactor. The amino acid differences lead to photochemical distinctions between PSII centers containing the two D1 forms (45, 53–55). We suspect these functional changes underlie the differential sensitivity of cells containing the two forms to UV-B, which is absorbed by PSII cofactors and D1 amino acid residues (2, 3, 50–52). An intrinsic difference in UV-B resistance between the forms appears likely, because the strain maintains a complex regulatory system to rapidly exchange the D1 forms under UV-B exposure. We have as yet no direct evidence for the mechanism of this differential susceptibility. At the 25 positions distinguishing D1:1 and D1:2 from Synechococcus, D1 from plants shares more common residues with D1:2 than with D1:1, including Glu130 as in D1:2. This raises the question of why Synechococcus maintains D1:1. We have some biophysical evidence that the D1:1 protein may prove superior under low and fluctuating light (53), which may account for the maintenance of two D1 forms over evolutionary time. Other workers have speculated that strains such as Synechococcus with phycocyanin-rich phycobilisomes may be more sensitive to UV-B than are strains with phycobilisomes containing phycoerythrin (19). In that case, the psbA exchange system might help compensate for UV-B absorbance by phycocyanin.
The exchange of alternate D1 forms does not completely account for UV effects on wild-type cells, because electron transport was more inhibited than PSII function alone. This pattern was also observed in the R2S2C3 strain and, to a lesser extent, in R2K1 (Fig. 7). We suspect that in R2K1, the high intrinsic resistance to UV-B inhibition of PSII function (Fig. 7A) allows the cells to minimize secondary effects of UV downstream from PSII and maintain electron transport near control rates. In contrast, the wild type is initially sensitive to UV-B inhibition of PSII (Fig. 7A), which may make the cells susceptible to downstream UV-B effects on electron transport (Fig. 7B), which then persist longer than the transient initial PSII inhibition. The electron transport inhibition may reflect UV-B effects outside PSII such as inhibition of rbcL expression or the activity of ribulose-1,5-bisphosphate carboxylase/oxygenase (9–12). Such downstream effects of UV-B could feedback to inhibit electron transport beyond PSII.
The psbA System May Prove Important in Community-Level Responses to UV-B.
The D1 exchange system accounts for much of this cyanobacterium’s capacity for photosynthetic acclimation to moderate UV-B exposure. This rapid response converts cells from a UV-B-susceptible to a UV-B-resistant phenotype within 15–30 min, and the protection is sustained for at least 2 h. All cyanobacteria examined to date contain psbA gene families and this prokaryotic isozyme system (47) may prove central to some community responses to UV-B exposure. Phytoplankton communities often remain fairly stable in the face of increasing UV-B (32, 35), but their resistance to UV-B inhibition of community-level photosynthesis varies strongly over the day (56), with maximal resistance to UV-B achieved around mid-day (4, 32). These field results are clear evidence for dynamic hour-scale acclimation processes that modulate community responses to UV-B. Key aspects of ecological modeling are the extent and limits of UV repair and adaptation processes in major biomass producers (56). The phenotypic plasticity imparted by the psbA system may underlie cyanobacterial community resilience in the face of moderate UV-B. This cyanobacterial response, however, is distinct from UV-B resistance mechanisms in organisms with chloroplasts, which contain only one active psbA gene (57, 58).
Acknowledgments
This work was supported by grants from the Swedish Natural Sciences Research Council (G.Ö., P.G., and A.K.C.) and the Seth M. Kempe Memorial Foundation (A.K.C.).
ABBREVIATIONS
- PSII
photosystem II
- Chl
chlorophyll
References
- 1.Aro E M, Virgin I, Andersson B. Biochim Biophys Act. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg B, Gaba V, Canaani O, Malkin S, Mattoo A K, Edelman M. Proc Natl Acad Sci USA. 1989;86:6617–6620. doi: 10.1073/pnas.86.17.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen M A K, Gaba V, Greenberg B, Mattoo A K, Edelman M. In: Photosynthetic Responses to the Environment. Yamamoto H Y, Smith C M, editors. Rockville, MD: Am. Soc. Plant Physiol.; 1993. pp. 142–149. [Google Scholar]
- 4.Prézelin B B, Boucher N P, Smith R C. In: Photosynthetic Responses to the Environment. Yamamoto H Y, Smith C M, editors. Rockville, MD: Am. Soc. Plant Physiol.; 1993. pp. 150–155. [Google Scholar]
- 5.Schofield O, Kroon B M A, Prezelin B B. J Phycol. 1995;31:703–715. [Google Scholar]
- 6.Nedunchezhian N, Kulandaivelu G. Acta Physiol Plant. 1996;18:39–45. [Google Scholar]
- 7.Rajagopal S, Murthy S D S. Photosynthetica. 1996;32:281–284. [Google Scholar]
- 8.Quesada A, Mouget J L, Vincent W F. J Phycol. 1995;31:242–248. [Google Scholar]
- 9.Jordan B R, He J, Chow W S, Anderson J M. Plant Cell Environ. 1991;15:91–98. [Google Scholar]
- 10.Mackerness S A H, Thomas B, Jordon B R. J Exp Bot. 1996;48:729–738. [Google Scholar]
- 11.Lesser M P. Mar Ecol Prog Ser. 1996;132:287–297. [Google Scholar]
- 12.Lao K, Glazer A N. Proc Natl Acad Sci USA. 1996;93:5258–5263. doi: 10.1073/pnas.93.11.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donkor V A, Hader D P. Act Protozoo. 1997;36:49–55. [Google Scholar]
- 14.Donkor V A, Hader D P. Aquat Microbiol Ecol. 1996;11:143–149. [Google Scholar]
- 15.Araoz R, Hader D P. FEMS Microbiol Ecol. 1997;23:301–313. [Google Scholar]
- 16.Sinha R P, Lebert M, Kumar A, Kumar H D, Hader D P. Bot Acta. 1995;108:87–92. [Google Scholar]
- 17.Kumar A, Sinha R P, Häder D-P. J Plant Physiol. 1996;148:86–91. [Google Scholar]
- 18.Sinha R P, Singh N, Kumar A, Kumar H D, Häder M, Häder D-P. J Photochem Photobiol. 1996;32:107–113. [Google Scholar]
- 19.Tyagi R, Srinivas G, Vyas D, Kumar A, Kumar H D. Photochem Photobiol. 1992;55:401–407. doi: 10.1111/j.1751-1097.1992.tb04254.x. [DOI] [PubMed] [Google Scholar]
- 20.Blakefield M K, Harris D O. Photochem Photobiol. 1994;59:204–208. doi: 10.1111/j.1751-1097.1994.tb05023.x. [DOI] [PubMed] [Google Scholar]
- 21.Fiscus E L, Booker F L. Photosynth Res. 1995;43:81–92. doi: 10.1007/BF00042965. [DOI] [PubMed] [Google Scholar]
- 22.Curtis S E, Haselkorn R. Plant Mol Biol. 1984;3:249–258. doi: 10.1007/BF00029661. [DOI] [PubMed] [Google Scholar]
- 23.Mulligan B, Schultes N, Chen L, Bogorad L. Proc Natl Acad Sci USA. 1984;81:2693–2697. doi: 10.1073/pnas.81.9.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golden S S, Brusslan J, Haselkorn R. EMBO J. 1986;5:2789–2798. doi: 10.1002/j.1460-2075.1986.tb04569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansson C, Debus R J, Osiewacz H D, Gurevitz M, McIntosh L. Plant Physiol. 1987;85:1021–1025. doi: 10.1104/pp.85.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaefer M R, Golden S S. J Biol Chem. 1989;264:7412–7417. [PubMed] [Google Scholar]
- 27.Clarke A K, Soitamo A, Gustafsson P, Öquist G. Proc Natl Acad Sci USA. 1993;90:9973–9977. doi: 10.1073/pnas.90.21.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulkarni R D, Golden S S. J Bacteriol. 1994;176:959–965. doi: 10.1128/jb.176.4.959-965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarke A K, Campbell D, Gustafsson P, Öquist G. Planta. 1995;197:553–562. [Google Scholar]
- 30.Campbell D, Zhou G, Gustafsson P, Öquist G, Clarke A K. EMBO J. 1995;14:5457–5466. doi: 10.1002/j.1460-2075.1995.tb00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanier R Y, Cohen-Bazire G. Annu Rev Microbiol. 1977;31:225–274. doi: 10.1146/annurev.mi.31.100177.001301. [DOI] [PubMed] [Google Scholar]
- 32.Cullen J J, Neale P J. In: Photosynthetic Responses to the Environment. Yamamoto H Y, Smith C M, editors. Rockville, MD: Am. Soc. Plant Physiol.; 1993. pp. 45–60. [Google Scholar]
- 33.Cullen J J, Neale P J. Photosynth Res. 1994;39:303–320. doi: 10.1007/BF00014589. [DOI] [PubMed] [Google Scholar]
- 34.Schindler D W, Curtis P J, Parker B R, Stainton M P. Nature (London) 1996;379:705–707. [Google Scholar]
- 35.Vincent W F, Roy S. Environ Rev. 1993;1:1–12. [Google Scholar]
- 36.Lesser M P, Cullen J J, Neale P J. J Phycol. 1994;30:183–192. [Google Scholar]
- 37.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. J Gen Microbiol. 1989;111:1–61. [Google Scholar]
- 38.Myers J, Graham J R, Wang R T. Plant Physiol. 1980;66:1144–1149. doi: 10.1104/pp.66.6.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 40.Kulkarni R D, Schaefer M R, Golden S S. J Bacteriol. 1992;174:3775–3781. doi: 10.1128/jb.174.11.3775-3781.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clarke A K, Critchley C. Plant Physiol. 1992;100:2081–2089. doi: 10.1104/pp.100.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhalerao R P, Lind L K, Gustafsson P. Plant Mol Biol. 1994;26:313–326. doi: 10.1007/BF00039542. [DOI] [PubMed] [Google Scholar]
- 43.van Kooten O, Snel J F H. Photosynth Res. 1990;25:147–150. doi: 10.1007/BF00033156. [DOI] [PubMed] [Google Scholar]
- 44.Bustos S A, Schaefer M R, Golden S S. J Bacteriol. 1990;172:1998–2004. doi: 10.1128/jb.172.4.1998-2004.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clarke A K, Hurry V M, Gustafsson P, Öquist G. Proc Natl Acad Sci USA. 1993;90:12985–12989. doi: 10.1073/pnas.90.24.11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panagopoulos I, Bornman J F, Björn L O. Physiol Plant. 1992;84:140–145. [Google Scholar]
- 47.Golden S S. J Bacteriol. 1995;177:1651–1654. doi: 10.1128/jb.177.7.1651-1654.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bustos S A, Golden S S. Mol Gen Genet. 1992;232:221–230. doi: 10.1007/BF00280000. [DOI] [PubMed] [Google Scholar]
- 49.Tsinoremas N F, Schaefer M R, Golden S S. J Biol Chem. 1994;269:16143–16147. [PubMed] [Google Scholar]
- 50.Barbato R, Frizzo A, Friso G, Rigoni F, Giacometti C M. Eur J Biochem. 1995;227:723–729. doi: 10.1111/j.1432-1033.1995.tb20194.x. [DOI] [PubMed] [Google Scholar]
- 51.Tandori J, Mate Z, Maroti P, Vass I. Photosynth Res. 1996;50:171–179. doi: 10.1007/BF00014887. [DOI] [PubMed] [Google Scholar]
- 52.Spetea C, Hideg E, Vass I. Plant Sci. 1996;115:207–215. [Google Scholar]
- 53.Campbell D, Bruce D, Carpenter C, Gustafsson P, Öquist G. Photosynth Res. 1996;47:131–144. doi: 10.1007/BF00016176. [DOI] [PubMed] [Google Scholar]
- 54.Krupa Z, Öquist G, Gustafsson P. Physiol Plant. 1991;82:1–8. doi: 10.1104/pp.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giorgi L B, Nixon P J, Merry S A P, Joseph D M, Durrant J R, Rivas J D, Barber J, Porter G, Klug D R. J Biol Chem. 1996;2696:2093–2101. doi: 10.1074/jbc.271.4.2093. [DOI] [PubMed] [Google Scholar]
- 56.Hader D P. Plant Ecol. 1997;128:4–13. [Google Scholar]
- 57.Jansen M A K, Babu T S, Heller D, Gaba V, Mattoo A K, Edelman M. Plant Sci. 1996;115:217–223. [Google Scholar]
- 58.Nilawati J, Greenberg B M, Smith R E H. J Phycol. 1997;33:215–224. [Google Scholar]