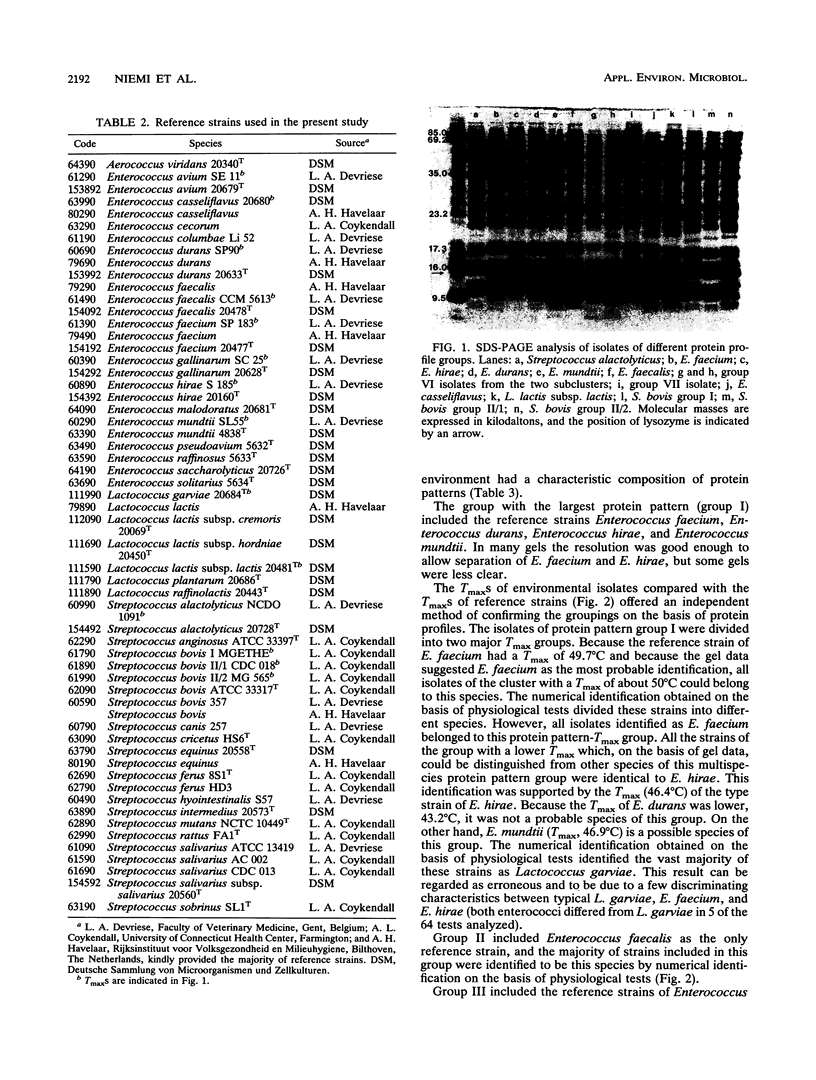
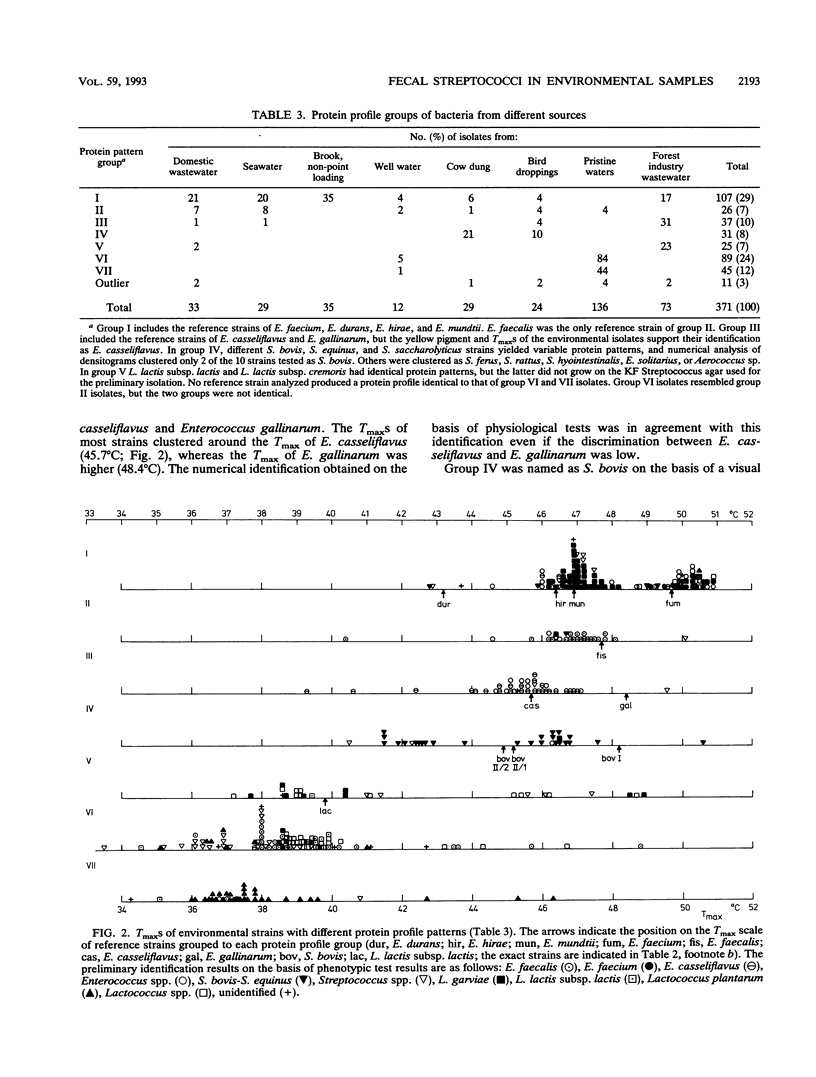
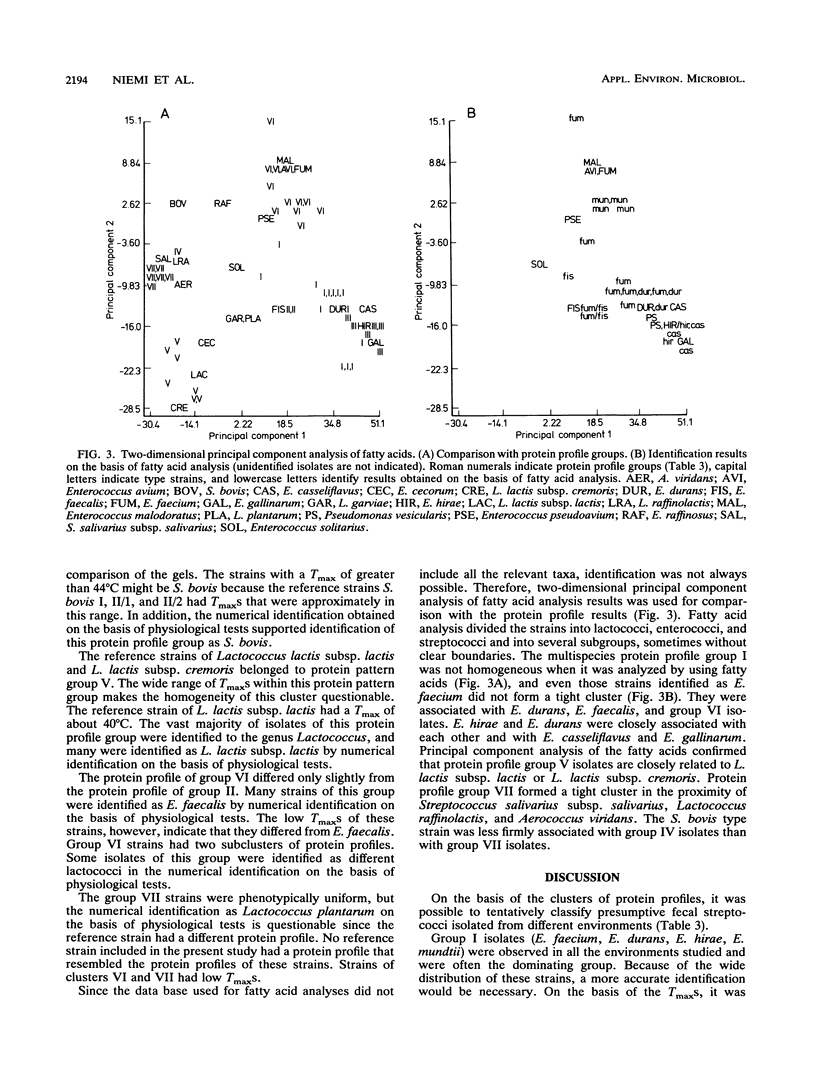
Abstract
The use of fecal streptococci as fecal indicators requires better knowledge of the ecology of these bacteria. We isolated 371 presumptive fecal streptococci from environmental samples--domestic wastewater, forest industry wastewater, contaminated surface and seawater, well water, cow dung, bird droppings, and pristine waters--and clustered them according to their protein profiles in one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. Some clusters could be tentatively identified with the help of reference strains. Samples from each environment had a typical composition of streptococcus types. Enterococcus faecalis was present, but not as a dominating enterococcal species, in samples in which fecal contamination was probable. Enterococcus faecium, Enterococcus durans, Enterococcus hirae, and Enterococcus mundtii had protein profiles that were difficult to distinguish from each other. These bacteria were found in a variety of samples. Enterococcus casseliflavus and Enterococcus gallinarum had identical protein profiles. On the basis of the maximum temperatures for growth and pigment production, isolates of this protein profile group common in forest industry wastewaters were identified as E. casseliflavus. Lactococcus lactis subsp. lactis was also found in this environment. Nearly all strains from pristine waters belonged to protein profile groups which could not be identified with the aid of known Aerococcus, Enterococcus, Lactococcus, or Streptococcus strains. The maximum temperatures for growth and the results of fatty acid analysis were in general agreement within each protein profile group.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cabelli V. J., Dufour A. P., McCabe L. J., Levin M. A. Swimming-associated gastroenteritis and water quality. Am J Epidemiol. 1982 Apr;115(4):606–616. doi: 10.1093/oxfordjournals.aje.a113342. [DOI] [PubMed] [Google Scholar]
- Dybowski W., Franklin D. A. Conditional probability and the identification of bacteria: a pilot study. J Gen Microbiol. 1968 Dec;54(2):215–229. doi: 10.1099/00221287-54-2-215. [DOI] [PubMed] [Google Scholar]
- Ferley J. P., Zmirou D., Balducci F., Baleux B., Fera P., Larbaigt G., Jacq E., Moissonnier B., Blineau A., Boudot J. Epidemiological significance of microbiological pollution criteria for river recreational waters. Int J Epidemiol. 1989 Mar;18(1):198–205. doi: 10.1093/ije/18.1.198. [DOI] [PubMed] [Google Scholar]
- Hantula J., Korhonen T. K., Bamford D. H. Determination of taxonomic resolution capacity of conventional one-dimensional SDS-polyacrylamide gel electrophoresis of whole-cell proteins using Enterobacteriaceae. FEMS Microbiol Lett. 1990 Aug;58(3):325–330. doi: 10.1111/j.1574-6968.1990.tb13998.x. [DOI] [PubMed] [Google Scholar]
- Kersters K., De Ley J. Classification and identification of bacteria by electrophoresis of their proteins. Soc Appl Bacteriol Symp Ser. 1980;8:273–297. [PubMed] [Google Scholar]
- Olkkonen V. M., Bamford D. H. Quantitation of the adsorption and penetration stages of bacteriophage phi 6 infection. Virology. 1989 Jul;171(1):229–238. doi: 10.1016/0042-6822(89)90530-8. [DOI] [PubMed] [Google Scholar]
- SWAN A. The use of a bile-aesculin medium and of Maxted's technique of Lancefield grouping in the identification of enterococci (group D streptococci). J Clin Pathol. 1954 May;7(2):160–163. doi: 10.1136/jcp.7.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox W. R., Lapage S. P., Bascomb S., Curtis M. A. Identification of bacteria by computer: theory and programming. J Gen Microbiol. 1973 Aug;77(2):317–330. doi: 10.1099/00221287-77-2-317. [DOI] [PubMed] [Google Scholar]
- Williams A. M., Rodrigues U. M., Collins M. D. Intrageneric relationships of Enterococci as determined by reverse transcriptase sequencing of small-subunit rRNA. Res Microbiol. 1991 Jan;142(1):67–74. doi: 10.1016/0923-2508(91)90098-u. [DOI] [PubMed] [Google Scholar]