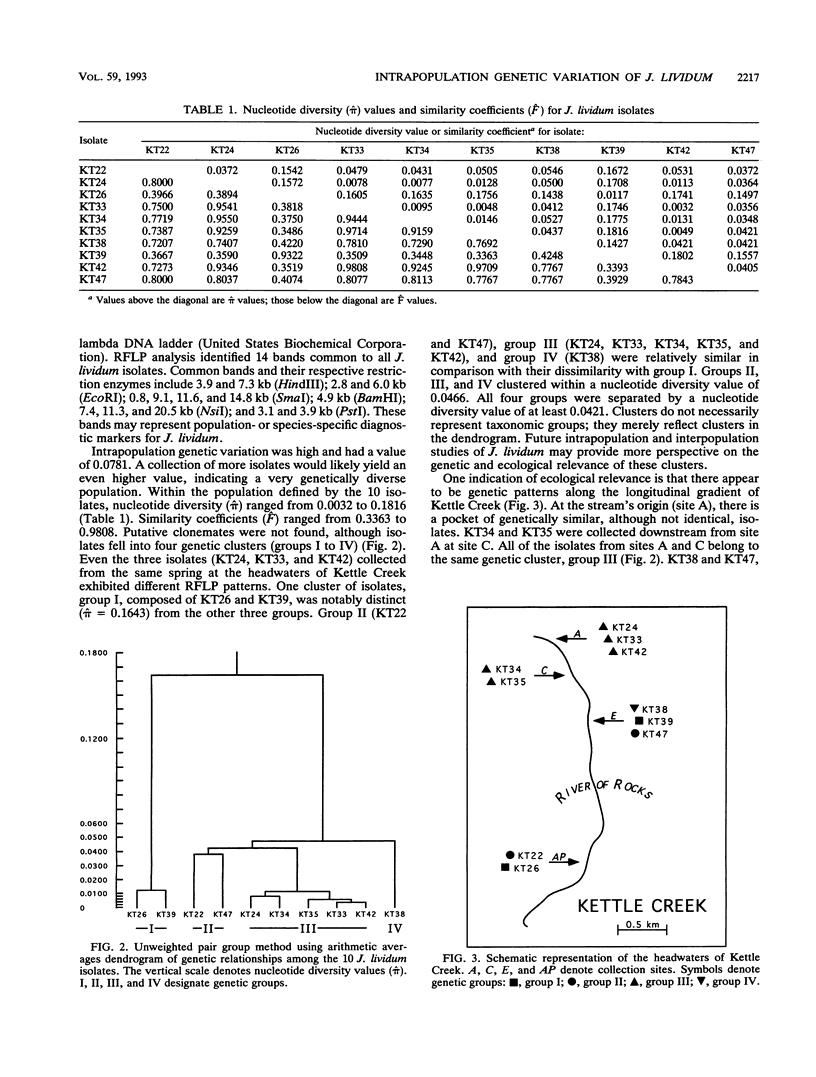
Abstract
An understanding of the genetic variation within and between populations should allow scientists to address many problems, including those associated with endangered species and the release of genetically modified organisms into the environment. With respect to microorganisms, the release of genetically engineered microorganisms is likely to increase dramatically given the current growth in the bioremediation industry. In this study, genetic variation within a lotic, bacterial population of Janthinobacterium lividum was measured with restriction fragment length polymorphism analysis. Chromosomal DNA from 10 Kettle Creek (Hawk Mountain Sanctuary, Kempton, Pa.) J. lividum isolates was digested with six restriction endonucleases and probed with a 7.5-kb pKK3535 fragment containing the E. coli rrnB rRNA operon. Genetic variation, as measured in terms of nucleotide diversity, was high within the population. The 0.0781 value for genetic variation was especially high given the conservative nature of the genetic probe. The average percent similarity among isolates within the population was 67.25%. Pairwise comparisons of nucleotide diversity values (π) and similarity coefficients (F) yielded values ranging from 0.0032 to 0.1816 and 0.3363 to 0.9808, respectively. Putative clonemates were not present within the group of isolates; however, all isolates shared 14 fragments across a spectrum of six restriction enzymes. The presence of these common fragments indicates that restriction fragment length polymorphism analysis may provide population- or species-specific diagnostic markers for J. lividum. Data that suggest a plume effect with respect to the downstream movement of J. lividum are also presented. An increase in genetic variation within groups of isolates along the longitudinal gradient of Kettle Creek is also suggested.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Brosius J., Ullrich A., Raker M. A., Gray A., Dull T. J., Gutell R. R., Noller H. F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981 Jul;6(1):112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- De Buyser M. L., Morvan A., Grimont F., el Solh N. Characterization of Staphylococcus species by ribosomal RNA gene restriction patterns. J Gen Microbiol. 1989 Apr;135(4):989–999. doi: 10.1099/00221287-135-4-989. [DOI] [PubMed] [Google Scholar]
- Eardly B. D., Materon L. A., Smith N. H., Johnson D. A., Rumbaugh M. D., Selander R. K. Genetic structure of natural populations of the nitrogen-fixing bacterium Rhizobium meliloti. Appl Environ Microbiol. 1990 Jan;56(1):187–194. doi: 10.1128/aem.56.1.187-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genthner F. J., Campbell R. P., Pritchard P. H. Use of a novel plasmid to monitor the fate of a genetically engineered Pseudomonas putida strain. Mol Ecol. 1992 Oct;1(3):137–143. doi: 10.1111/j.1365-294x.1992.tb00169.x. [DOI] [PubMed] [Google Scholar]
- Hadrys H., Balick M., Schierwater B. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol Ecol. 1992 May;1(1):55–63. doi: 10.1111/j.1365-294x.1992.tb00155.x. [DOI] [PubMed] [Google Scholar]
- Istock C. A., Duncan K. E., Ferguson N., Zhou X. Sexuality in a natural population of bacteria--Bacillus subtilis challenges the clonal paradigm. Mol Ecol. 1992 Aug;1(2):95–103. doi: 10.1111/j.1365-294x.1992.tb00161.x. [DOI] [PubMed] [Google Scholar]
- Masters C. I., Murray R. G., Moseley B. E., Minton K. W. DNA polymorphisms in new isolates of 'Deinococcus radiopugnans'. J Gen Microbiol. 1991 Jul;137(7):1459–1469. doi: 10.1099/00221287-137-7-1459. [DOI] [PubMed] [Google Scholar]
- McArthur J. V., Kovacic D. A., Smith M. H. Genetic diversity in natural populations of a soil bacterium across a landscape gradient. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9621–9624. doi: 10.1073/pnas.85.24.9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard-Pasquier N., Picard B., Heeralal S., Krishnamoorthy R., Goullet P. Correlation between ribosomal DNA polymorphism and electrophoretic enzyme polymorphism in Yersinia. J Gen Microbiol. 1990 Aug;136(8):1655–1666. doi: 10.1099/00221287-136-8-1655. [DOI] [PubMed] [Google Scholar]
- Rodrigues U. M., Aguirre M., Facklam R. R., Collins M. D. Specific and intraspecific molecular typing of lactococci based on polymorphism of DNA encoding rRNA. J Appl Bacteriol. 1991 Dec;71(6):509–516. doi: 10.1111/j.1365-2672.1991.tb03825.x. [DOI] [PubMed] [Google Scholar]
- Saunders N. A., Harrison T. G., Kachwalla N., Taylor A. G. Identification of species of the genus Legionella using a cloned rRNA gene from Legionella pneumophila. J Gen Microbiol. 1988 Aug;134(8):2363–2374. doi: 10.1099/00221287-134-8-2363. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short K. A., King R. J., Seidler R. J., Olsen R. H. Biodegradation of phenoxyacetic acid in soil by Pseudomonas putida PP0301(pR0103), a constitutive degrader of 2,4-dichlorophenoxyacetate. Mol Ecol. 1992 Aug;1(2):89–94. doi: 10.1111/j.1365-294x.1992.tb00160.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987 May 15;236(4803):787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Thomson-Carter F. M., Carter P. E., Pennington T. H. Differentiation of staphylococcal species and strains by ribosomal RNA gene restriction patterns. J Gen Microbiol. 1989 Jul;135(7):2093–2097. doi: 10.1099/00221287-135-7-2093. [DOI] [PubMed] [Google Scholar]
- Williams A. M., Collins M. D. DNA fingerprinting of Streptococcus uberis based on polymorphism of DNA encoding rRNA. Lett Appl Microbiol. 1991 Jan;12(1):23–28. doi: 10.1111/j.1472-765x.1991.tb00493.x. [DOI] [PubMed] [Google Scholar]
- Williams D. L., Gillis T. P., Portaels F. Geographically distinct isolates of Mycobacterium leprae exhibit no genotypic diversity by restriction fragment-length polymorphism analysis. Mol Microbiol. 1990 Oct;4(10):1653–1659. doi: 10.1111/j.1365-2958.1990.tb00542.x. [DOI] [PubMed] [Google Scholar]
- el-Adhami W., Roberts L., Vickery A., Inglis B., Gibbs A., Stewart P. R. Epidemiological analysis of a methicillin-resistant Staphylococcus aureus outbreak using restriction fragment length polymorphisms of genomic DNA. J Gen Microbiol. 1991 Dec;137(12):2713–2720. doi: 10.1099/00221287-137-12-2713. [DOI] [PubMed] [Google Scholar]


