Abstract
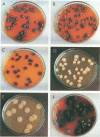
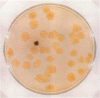
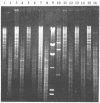
During a strain selection program to improve oxytetracycline production in Streptomyces rimosus R6, isolates that showed extreme morphological instability appeared. Propagation via spores gave much higher instability than did propagation via mycelial fragments. Five phenotypic traits were affected: sporulation, pigmentation, colony morphology, oxytetracycline production, and oxytetracycline resistance. The variants were classified on the basis of oxytetracycline resistance into three classes. Class I variants (99% of variants) showed parental levels of resistance but were very heterogeneous regarding the other phenotypes. No DNA rearrangements were detected in primary class I variants. Class II variants (1% of variants, oxytetracycline sensitive) were phenotypically uniform, and most variants carried the same large deletion of ca. 455 kb, including the oxytetracycline resistance gene otrB. Class III variants (0.1% of variants, increased oxytetracycline resistance) were phenotypically uniform and overproduced a brown pigment and oxytetracycline. Most of these variants also showed a reproducible large-scale DNA rearrangement, which probably included deletion and a low-level reiteration (three or four copies) of a DNA fragment. "Revertants" of some class I variants show a similar DNA rearrangement to the class III variants, but there is extensive reiteration of sequences of about 200 kb, including the otrB gene. The significance of these results for the problem of strain degeneration and overproduction of antibiotics is discussed.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenbuchner J., Cullum J. DNA amplification and an unstable arginine gene in Streptomyces lividans 66. Mol Gen Genet. 1984;195(1-2):134–138. doi: 10.1007/BF00332735. [DOI] [PubMed] [Google Scholar]
- Birch A., Häusler A., Vögtli M., Krek W., Hütter R. Extremely large chromosomal deletions are intimately involved in genetic instability and genomic rearrangements in Streptomyces glaucescens. Mol Gen Genet. 1989 Jun;217(2-3):447–458. doi: 10.1007/BF02464916. [DOI] [PubMed] [Google Scholar]
- Butler M. J., Friend E. J., Hunter I. S., Kaczmarek F. S., Sugden D. A., Warren M. Molecular cloning of resistance genes and architecture of a linked gene cluster involved in biosynthesis of oxytetracycline by Streptomyces rimosus. Mol Gen Genet. 1989 Jan;215(2):231–238. doi: 10.1007/BF00339722. [DOI] [PubMed] [Google Scholar]
- Cullum J., Altenbuchner J., Flett F., Piendl W. DNA amplification and genetic instability in Streptomyces. Biotechnol Genet Eng Rev. 1986;4:59–78. doi: 10.1080/02648725.1986.10647823. [DOI] [PubMed] [Google Scholar]
- Fishman S. E., Hershberger C. L. Amplified DNA in Streptomyces fradiae. J Bacteriol. 1983 Aug;155(2):459–466. doi: 10.1128/jb.155.2.459-466.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flett F., Cullum J. DNA deletions in spontaneous chloramphenicol-sensitive mutants of Streptomyces coelicolor A 3(2) and Streptomyces lividans 66. Mol Gen Genet. 1987 May;207(2-3):499–502. doi: 10.1007/BF00331621. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Hintermann G., Simonet J. M., Crameri R., Piret J., Hütter R. Certain chromosomal regions in Streptomyces glaucescens tend to carry amplifications and deletions. Mol Gen Genet. 1985;200(3):375–384. doi: 10.1007/BF00425720. [DOI] [PubMed] [Google Scholar]
- Hornemann U., Otto C. J., Hoffman G. G., Bertinuson A. C. Spectinomycin resistance and associated DNA amplification in Streptomyces achromogenes subsp. rubradiris. J Bacteriol. 1987 Jun;169(6):2360–2366. doi: 10.1128/jb.169.6.2360-2366.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hranueli D., Pigac J., Smokvina T., Alacević M. Genetic interactions in Streptomyces rimosus mediated by conjugation and by protoplast fusion. J Gen Microbiol. 1983 May;129(5):1415–1422. doi: 10.1099/00221287-129-5-1415. [DOI] [PubMed] [Google Scholar]
- Hranueli D., Pigac J., Vesligaj M. Characterization and persistence of actinophage RP2 isolated from Streptomyces rimosus ATCC 10970. J Gen Microbiol. 1979 Oct;114(2):295–303. doi: 10.1099/00221287-114-2-295. [DOI] [PubMed] [Google Scholar]
- Hranueli D., Smokvina T., Alacević M. A comparative study of protoplast preparation and regeneration in Streptomyces rimosus and Streptomyces lividans. Microbiologica. 1986 Jul;9(3):387–392. [PubMed] [Google Scholar]
- Häusler A., Birch A., Krek W., Piret J., Hütter R. Heterogeneous genomic amplification in Streptomyces glaucescens: structure, location and junction sequence analysis. Mol Gen Genet. 1989 Jun;217(2-3):437–446. doi: 10.1007/BF02464915. [DOI] [PubMed] [Google Scholar]
- Kaiser P., Flett F., Cullum J. Stabilization of Streptomyces lividans by homologous recombinational insertion. Biotechnology (N Y) 1992 May;10(5):570–573. doi: 10.1038/nbt0592-570. [DOI] [PubMed] [Google Scholar]
- Leblond P., Demuyter P., Simonet J. M., Decaris B. Genetic instability and hypervariability in Streptomyces ambofaciens: towards an understanding of a mechanism of genome plasticity. Mol Microbiol. 1990 May;4(5):707–714. doi: 10.1111/j.1365-2958.1990.tb00641.x. [DOI] [PubMed] [Google Scholar]
- Miyoshi T., Iseki M., Konomi T., Imanaka H. Biosynthesis of bicyclomycin. I. Appearance of aerial mycelia negative strains (am-). J Antibiot (Tokyo) 1980 May;33(5):480–487. doi: 10.7164/antibiotics.33.480. [DOI] [PubMed] [Google Scholar]
- Pigac J., Hranueli D., Smokvina T., Alacević M. Optimal Cultural and Physiological Conditions for Handling Streptomyces rimosus Protoplasts. Appl Environ Microbiol. 1982 Nov;44(5):1178–1186. doi: 10.1128/aem.44.5.1178-1186.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potekhin Ia A., Danilenko V. N. Determinant ustoichivosti k kanamitsinu Streptomyces rimosus: amplifikatsiia v sostave khromosomy i obratimaia geneticheskaia nestabil'nost'. Mol Biol (Mosk) 1985 May-Jun;19(3):805–817. [PubMed] [Google Scholar]
- Reynes J. P., Calmels T., Drocourt D., Tiraby G. Cloning, expression in Escherichia coli and nucleotide sequence of a tetracycline-resistance gene from Streptomyces rimosus. J Gen Microbiol. 1988 Mar;134(3):585–598. doi: 10.1099/00221287-134-3-585. [DOI] [PubMed] [Google Scholar]
- Rhodes P. M., Hunter I. S., Friend E. J., Warren M. Recombinant DNA methods for the oxytetracycline producer Streptomyces rimosus. Biochem Soc Trans. 1984 Aug;12(4):586–587. doi: 10.1042/bst0120586. [DOI] [PubMed] [Google Scholar]
- Schrempf H. Deletion and amplification of DNA sequences in melanin-negative variants of Streptomyces reticuli. Mol Gen Genet. 1983;189(3):501–505. doi: 10.1007/BF00325917. [DOI] [PubMed] [Google Scholar]
- Starodubtseva L. I., Taisova A. S., Danilenko V. N. Izuchenie iavleniia amplifikatsii determinanta ustoichivosti k kanamitsinu (Kanr) v sostave skonstruirovannykh gibridnykh plazmid v shtamme Streptomyces lividans. Antibiot Med Biotekhnol. 1985 Aug;30(8):565–572. [PubMed] [Google Scholar]