Abstract
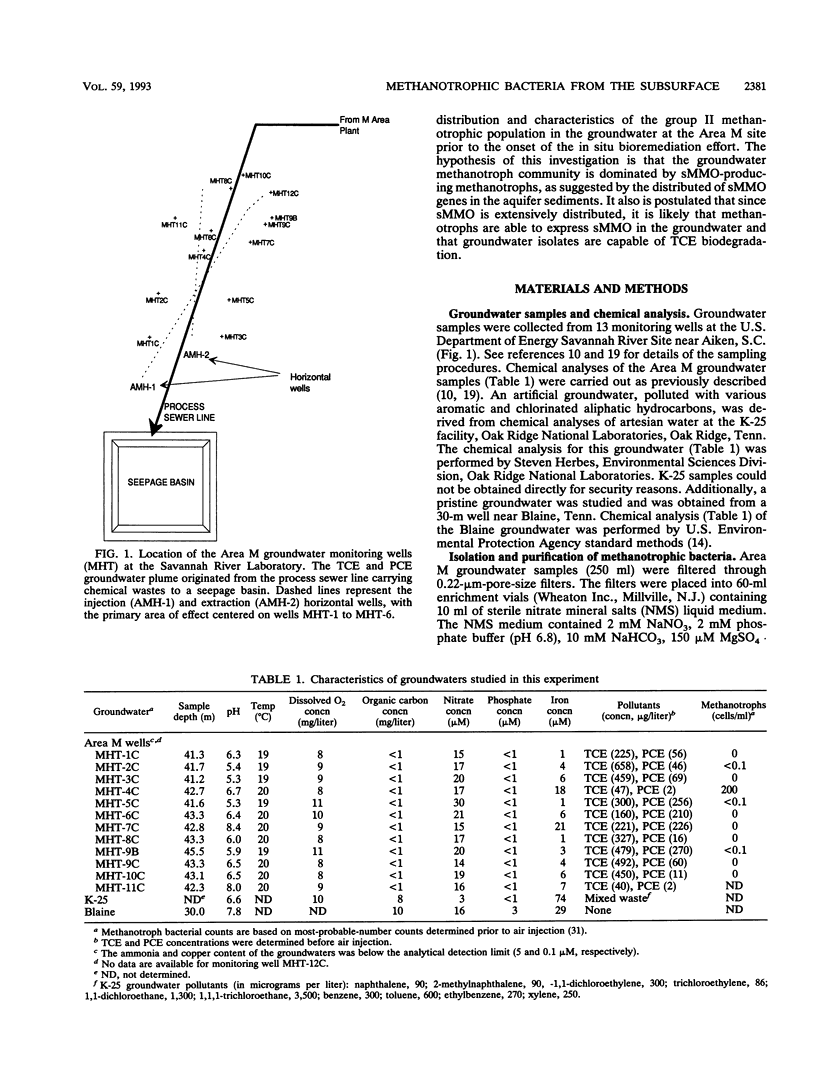
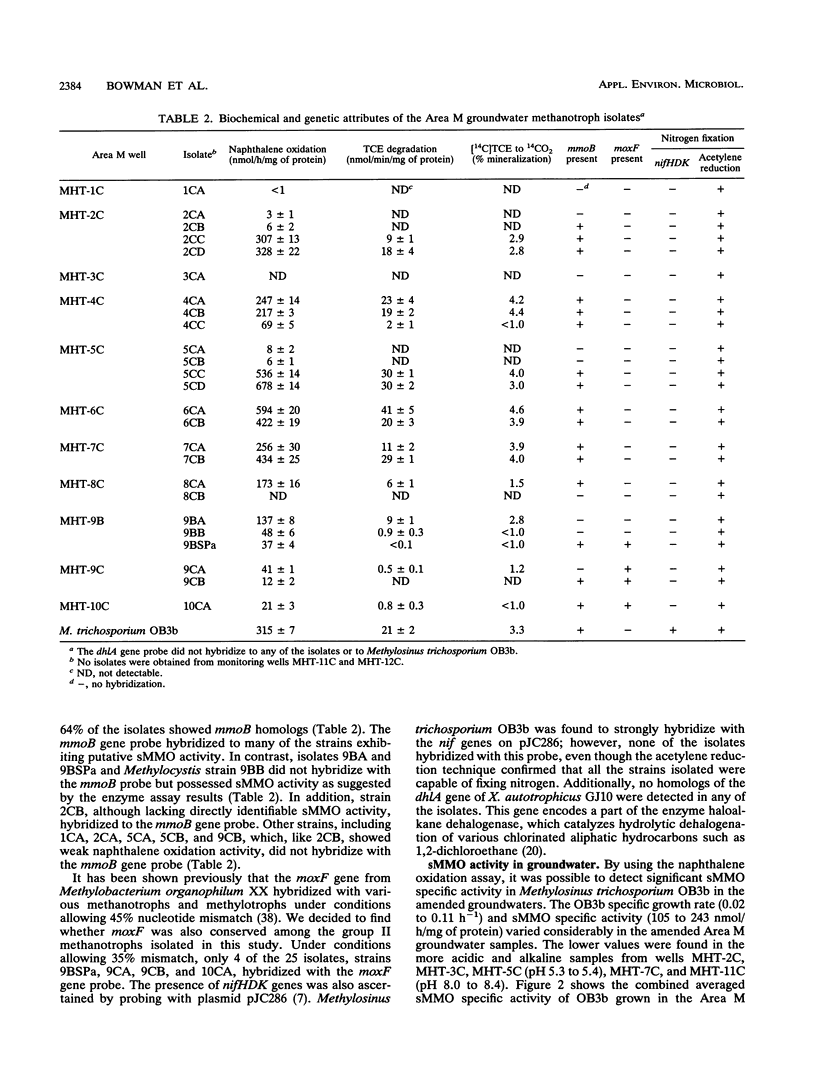
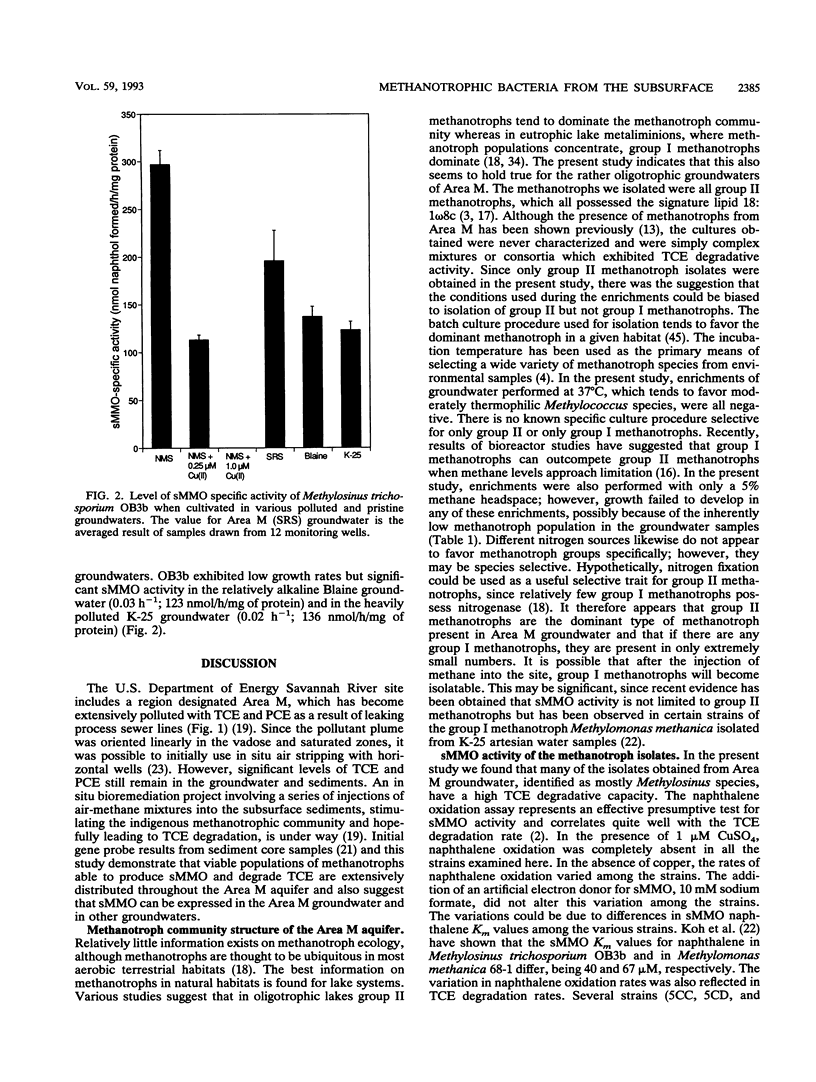
Groundwater, contaminated with trichloroethylene (TCE) and tetrachloroethylene (PCE), was collected from 13 monitoring wells at Area M on the U.S. Department of Energy Savannah River Site near Aiken, S.C. Filtered groundwater samples were enriched with methane, leading to the isolation of 25 methanotrophic isolates. The phospholipid fatty acid profiles of all the isolates were dominated by 18:1 omega 8c (60 to 80%), a signature lipid for group II methanotrophs. Subsequent phenotypic testing showed that most of the strains were members of the genus Methylosinus and one isolate was a member of the genus Methylocystis. Most of the methanotroph isolates exhibited soluble methane monooxygenase (sMMO) activity. This was presumptively indicated by the naphthalene oxidation assay and confirmed by hybridization with a gene probe encoding the mmoB gene and by cell extract assays. TCE was degraded at various rates by most of the sMMO-producing isolates, whereas PCE was not degraded. Savannah River Area M and other groundwaters, pristine and polluted, were found to support sMMO activity when supplemented with nutrients and then inoculated with Methylosinus trichosporium OB3b. The maximal sMMO-specific activity obtained in the various groundwaters ranged from 41 to 67% compared with maximal rates obtained in copper-free nitrate mineral salts media. This study partially supports the hypothesis that stimulation of indigenous methanotrophic communities can be efficacious for removal of chlorinated aliphatic hydrocarbons from subsurface sites and that the removal can be mediated by sMMO.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balkwill D. L., Fredrickson J. K., Thomas J. M. Vertical and horizontal variations in the physiological diversity of the aerobic chemoheterotrophic bacterial microflora in deep southeast coastal plain subsurface sediments. Appl Environ Microbiol. 1989 May;55(5):1058–1065. doi: 10.1128/aem.55.5.1058-1065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardy D. L., Laidler V., Salmond G. P., Murrell J. C. Molecular analysis of the methane monooxygenase (MMO) gene cluster of Methylosinus trichosporium OB3b. Mol Microbiol. 1991 Feb;5(2):335–342. doi: 10.1111/j.1365-2958.1991.tb02114.x. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. J., Brill W. J. Control of Klebsiella pneumoniae nif mRNA synthesis. J Bacteriol. 1985 Jun;162(3):1186–1190. doi: 10.1128/jb.162.3.1186-1190.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensley B. D. Biochemical diversity of trichloroethylene metabolism. Annu Rev Microbiol. 1991;45:283–299. doi: 10.1146/annurev.mi.45.100191.001435. [DOI] [PubMed] [Google Scholar]
- Fliermans C. B., Phelps T. J., Ringelberg D., Mikell A. T., White D. C. Mineralization of trichloroethylene by heterotrophic enrichment cultures. Appl Environ Microbiol. 1988 Jul;54(7):1709–1714. doi: 10.2172/666263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guckert J. B., Ringelberg D. B., White D. C., Hanson R. S., Bratina B. J. Membrane fatty acids as phenotypic markers in the polyphasic taxonomy of methylotrophs within the Proteobacteria. J Gen Microbiol. 1991 Nov;137(11):2631–2641. doi: 10.1099/00221287-137-11-2631. [DOI] [PubMed] [Google Scholar]
- Hanson R. S., Wattenberg E. V. Ecology of methylotrophic bacteria. Biotechnology. 1991;18:325–348. doi: 10.1016/b978-0-7506-9188-8.50021-8. [DOI] [PubMed] [Google Scholar]
- Janssen D. B., Pries F., van der Ploeg J., Kazemier B., Terpstra P., Witholt B. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J Bacteriol. 1989 Dec;171(12):6791–6799. doi: 10.1128/jb.171.12.6791-6799.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S. C., Bowman J. P., Sayler G. S. Soluble Methane Monooxygenase Production and Trichloroethylene Degradation by a Type I Methanotroph, Methylomonas methanica 68-1. Appl Environ Microbiol. 1993 Apr;59(4):960–967. doi: 10.1128/aem.59.4.960-967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUNKRES K. D., RICHARDS F. M. THE PURIFICATION AND PROPERTIES OF NEUROSPORA MALATE DEHYDROGENASE. Arch Biochem Biophys. 1965 Mar;109:466–479. doi: 10.1016/0003-9861(65)90391-7. [DOI] [PubMed] [Google Scholar]
- Machlin S. M., Hanson R. S. Nucleotide sequence and transcriptional start site of the Methylobacterium organophilum XX methanol dehydrogenase structural gene. J Bacteriol. 1988 Oct;170(10):4739–4747. doi: 10.1128/jb.170.10.4739-4747.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenhuis R., Oedzes J. Y., van der Waarde J. J., Janssen D. B. Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl Environ Microbiol. 1991 Jan;57(1):7–14. doi: 10.1128/aem.57.1.7-14.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayler G. S., Shields M. S., Tedford E. T., Breen A., Hooper S. W., Sirotkin K. M., Davis J. W. Application of DNA-DNA colony hybridization to the detection of catabolic genotypes in environmental samples. Appl Environ Microbiol. 1985 May;49(5):1295–1303. doi: 10.1128/aem.49.5.1295-1303.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R. L., Haygood M. G., Lidstrom M. E. Identification of putative methanol dehydrogenase (moxF) structural genes in methylotrophs and cloning of moxF genes from Methylococcus capsulatus bath and Methylomonas albus BG8. J Bacteriol. 1988 May;170(5):2063–2069. doi: 10.1128/jb.170.5.2063-2069.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Brusseau G. A., Hanson R. S., Waclett L. P. Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1989 Dec;55(12):3155–3161. doi: 10.1128/aem.55.12.3155-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Hanson R. S. Soluble methane monooxygenase component B gene probe for identification of methanotrophs that rapidly degrade trichloroethylene. Appl Environ Microbiol. 1992 Mar;58(3):953–960. doi: 10.1128/aem.58.3.953-960.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama H., Nakajima T., Yagi O., Nakahara T. Role of heterotrophic bacteria in complete mineralization of trichloroethylene by Methylocystis sp. strain M. Appl Environ Microbiol. 1992 Sep;58(9):3067–3071. doi: 10.1128/aem.58.9.3067-3071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett L. P., Gibson D. T. Rapid method for detection and quantitation of hydroxylated aromatic intermediates produced by microorganisms. Appl Environ Microbiol. 1983 Mar;45(3):1144–1147. doi: 10.1128/aem.45.3.1144-1147.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittenbury R., Davies S. L., Davey J. F. Exospores and cysts formed by methane-utilizing bacteria. J Gen Microbiol. 1970 May;61(2):219–226. doi: 10.1099/00221287-61-2-219. [DOI] [PubMed] [Google Scholar]



