Abstract
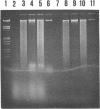
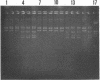
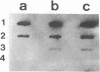
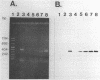
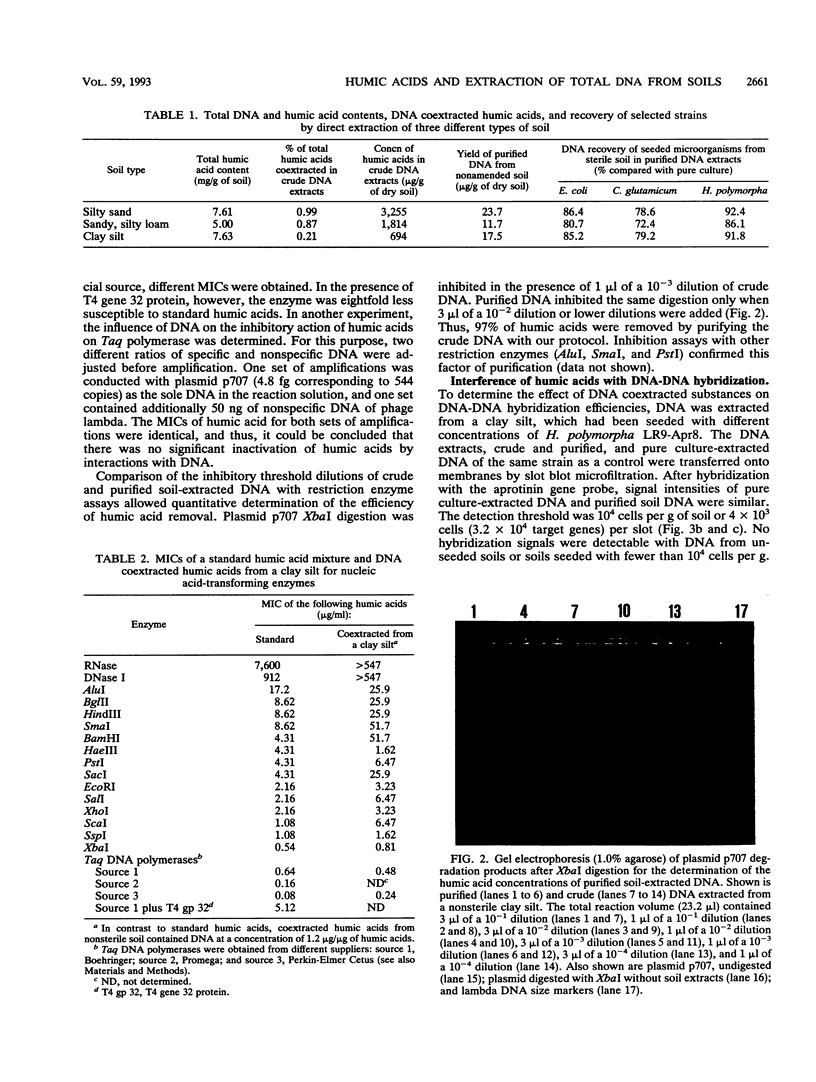
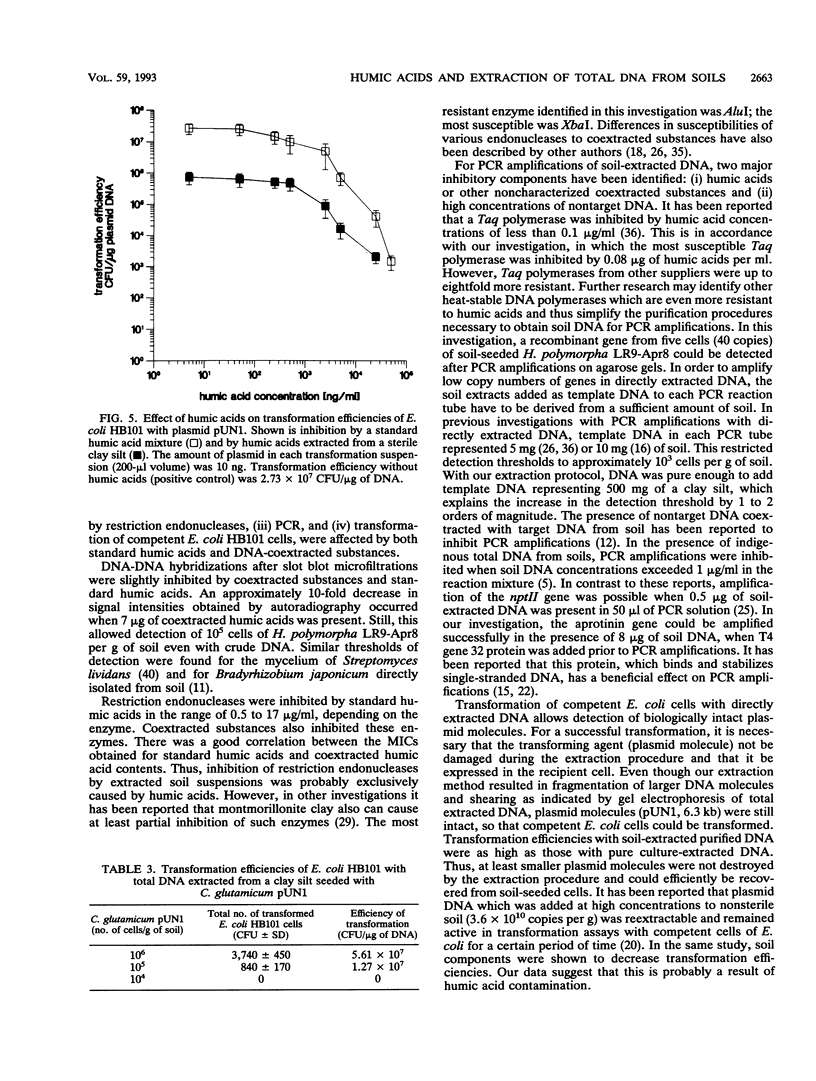
A two-step protocol for the extraction and purification of total DNA from soil samples was developed. Crude DNA extracts (100 microliters from 5 g of soil) were contaminated with humic acids at concentrations of 0.7 to 3.3 micrograms/microliters, depending on the type of soil extracted. The coextracted humic acid fraction of a clay silt was similar to a commercially available standard humic acid mixture, as determined by electrophoretic mobility in agarose gels, UV fluorescence, and inhibition assays with DNA-transforming enzymes. Restriction endonucleases were inhibited at humic acid concentrations of 0.5 to 17.2 micrograms/ml for the commercial product and 0.8 to 51.7 micrograms/ml for the coextracted humic acids. DNase I was less susceptible (MIC of standard humic acids, 912 micrograms/ml), and RNase could not be inhibited at all (MIC, > 7.6 mg/ml). High inhibitory susceptibilities for humic acids were observed with Taq polymerase. For three Taq polymerases from different commercial sources, MICs were 0.08 to 0.64 micrograms of the standard humic acids per ml and 0.24 to 0.48 micrograms of the coextracted humic acids per ml. The addition of T4 gene 32 protein increased the MIC for one Taq polymerase to 5.12 micrograms/ml. Humic acids decreased nonradioactive detection in DNA-DNA slot blot hybridizations at amounts of 0.1 micrograms and inhibited transformation of competent Escherichia coli HB101 with a broad-host-range plasmid, pUN1, at concentrations of 100 micrograms/ml. Purification of crude DNA with ion-exchange chromatography resulted in removal of 97% of the initially coextracted humic acids.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brake A. J., Merryweather J. P., Coit D. G., Heberlein U. A., Masiarz F. R., Mullenbach G. T., Urdea M. S., Valenzuela P., Barr P. J. Alpha-factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4642–4646. doi: 10.1073/pnas.81.15.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce K. D., Hiorns W. D., Hobman J. L., Osborn A. M., Strike P., Ritchie D. A. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl Environ Microbiol. 1992 Oct;58(10):3413–3416. doi: 10.1128/aem.58.10.3413-3416.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R. C., Shultz J. L., Lehman D. J. Alpha-factor leader sequence-directed transport of Escherichia coli beta-galactosidase in the secretory pathway of Saccharomyces cerevisiae. Mol Gen Genet. 1989 Aug;218(2):240–248. doi: 10.1007/BF00331274. [DOI] [PubMed] [Google Scholar]
- Eikmanns B. J., Kleinertz E., Liebl W., Sahm H. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene. 1991 Jun 15;102(1):93–98. doi: 10.1016/0378-1119(91)90545-m. [DOI] [PubMed] [Google Scholar]
- Fritz H., Wunderer G. Biochemistry and applications of aprotinin, the kallikrein inhibitor from bovine organs. Arzneimittelforschung. 1983;33(4):479–494. [PubMed] [Google Scholar]
- Gellissen G., Janowicz Z. A., Weydemann U., Melber K., Strasser A. W., Hollenberg C. P. High-level expression of foreign genes in Hansenula polymorpha. Biotechnol Adv. 1992;10(2):179–189. doi: 10.1016/0734-9750(92)90002-q. [DOI] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen C. S., Rasmussen O. F. Development and application of a new method to extract bacterial DNA from soil based on separation of bacteria from soil with cation-exchange resin. Appl Environ Microbiol. 1992 Aug;58(8):2458–2462. doi: 10.1128/aem.58.8.2458-2462.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan M. R., Oriel P. Transfer of Transposon Tn916 from Bacillus subtilis into a Natural Soil Population. Appl Environ Microbiol. 1992 Aug;58(8):2701–2703. doi: 10.1128/aem.58.8.2701-2703.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaccio M., Lew A. PCR based diagnosis in the presence of 8% (v/v) blood. Nucleic Acids Res. 1991 Mar 11;19(5):1151–1151. doi: 10.1093/nar/19.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard C., Ponsonnet C., Paget E., Nesme X., Simonet P. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1992 Sep;58(9):2717–2722. doi: 10.1128/aem.58.9.2717-2722.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S. D., Josephson K. L., Bailey R. L., Gerba C. P., Pepper I. L. Rapid method for processing soil samples for polymerase chain reaction amplification of specific gene sequences. Appl Environ Microbiol. 1991 Aug;57(8):2283–2286. doi: 10.1128/aem.57.8.2283-2286.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Sayler G., Wackernagel W. Persistence of free plasmid DNA in soil monitored by various methods, including a transformation assay. Appl Environ Microbiol. 1992 Sep;58(9):3012–3019. doi: 10.1128/aem.58.9.3012-3019.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K., Hansen-Hagge T., Bartram C. Improved yields of long PCR products using gene 32 protein. Nucleic Acids Res. 1990 Feb 25;18(4):1079–1079. doi: 10.1093/nar/18.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenska S., Klingmüller W. DNA recovery and direct detection of Tn5 sequences from soil. Lett Appl Microbiol. 1991 Jul;13(1):21–24. doi: 10.1111/j.1472-765x.1991.tb00559.x. [DOI] [PubMed] [Google Scholar]
- Selenska S., Klingmüller W. Direct recovery and molecular analysis of DNA and RNA from soil. Microb Releases. 1992 Jun;1(1):41–46. [PubMed] [Google Scholar]
- Smit E., van Elsas J. D., van Veen J. A., de Vos W. M. Detection of Plasmid Transfer from Pseudomonas fluorescens to Indigenous Bacteria in Soil by Using Bacteriophage phiR2f for Donor Counterselection. Appl Environ Microbiol. 1991 Dec;57(12):3482–3488. doi: 10.1128/aem.57.12.3482-3488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Goksøyr J., Bej A. K., Atlas R. M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988 Dec;54(12):2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V., Goksøyr J., Daae F. L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990 Mar;56(3):782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992 Feb;58(2):754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991 Apr;57(4):1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol. 1992 Jul;58(7):2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. G., Evans E. A., Blobel G. Prepro-alpha-factor has a cleavable signal sequence. J Biol Chem. 1988 May 5;263(13):6209–6214. [PubMed] [Google Scholar]
- Wipat A., Wellington E. M., Saunders V. A. Streptomyces marker plasmids for monitoring survival and spread of streptomycetes in soil. Appl Environ Microbiol. 1991 Nov;57(11):3322–3330. doi: 10.1128/aem.57.11.3322-3330.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]