Abstract
Protein E, a 91-residue membrane protein of φX174, causes lysis of the host in a growth-dependent manner reminiscent of cell wall antibiotics, suggesting E acts by inhibiting peptidoglycan synthesis. In a search for the cellular target of E, we previously have isolated recessive mutations in the host gene slyD (sensitivity to lysis) that block the lytic effects of E. The role of slyD, which encodes a FK506 binding protein-type peptidyl-prolyl cis-trans isomerase, is not fully understood. However, E mutants referred to as Epos (plates on slyD) lack a slyD requirement, indicating that slyD is not crucial for lysis. To identify the gene encoding the cellular target, we selected for survivors of Epos. In this study, we describe the isolation of dominant mutations in the essential host gene mraY that result in a general lysis-defective phenotype. mraY encodes translocase I, which catalyzes the formation of the first lipid-linked intermediate in cell wall biosynthesis. The isolation of these lysis-defective mutants supports a model in which translocase I is the cellular target of E and that inhibition of cell wall synthesis is the mechanism of lysis.
Most bacteriophages achieve release of progeny virions by causing lysis of the host cell. Double-stranded DNA phages generally encode at least two genes to elicit host cell lysis: an endolysin, which is an enzyme that degrades the cell wall, and a holin, which at a genetically defined time allows endolysin to cross the cytoplasmic membrane and gain access to the cell wall (1). Once endolysin degrades the cell wall, the cell lyses because of its internal osmotic pressure. In contrast, the small single-stranded DNA phage φX174 has a single lysis gene E of 91 codons (2). Expression of E from a multicopy plasmid is sufficient to cause lysis of Escherichia coli (3, 4). Gene fusion analysis has indicated that only the amino-terminal 35 aa of the E polypeptide, containing a hydrophobic region thought to be a transmembrane domain (TMD), is required for lytic activity (6, 7). The E protein has no detectable cell wall degrading activity, and given its simple primary structure, it is unlikely to have any enzymatic activity at all. Therefore, it must be promoting cell lysis by a mechanism distinct from direct degradation of the cell wall.
The mechanism by which E causes host cell lysis is controversial. Many models have been proposed to explain its lytic function. E-mediated lysis is strikingly similar to that caused by the antibiotic penicillin. Penicillin causes cell lysis by inhibiting cell wall synthesis at the level of peptide cross-link formation between strands of peptidoglycan. Both E and penicillin require cell growth for lysis, and light and electron microscopy have revealed that both result in lesions at the cell septum as intermediates in lysis (8–11). These observations led to a model in which E targets and inhibits a component of the cell wall synthesis pathway to cause cell lysis. However, it also has been reported that E-mediated lysis of hosts deficient in autolytic enzymes is impaired, leading to the hypothesis that E functions by targeting and activating a component of the E. coli autolytic system (12). Moreover, in scanning electron micrographs of cells undergoing E-mediated lysis, Lubitz and coworkers (13) have observed discrete 50- to 200-nm holes located at the cell septum and occasionally at the poles. This observation has led them to propose a model in which the E protein oligomerizes to form a “transmembrane tunnel” spanning the entire cell envelope to release the cytoplasmic contents, including progeny virions (13).
The confused picture for the mechanism of E lysis primarily is caused by the lack of molecular or genetic evidence to support any single model. As a consequence, we have taken a genetic approach to identify host genes required in E-mediated lysis. We previously have isolated recessive mutations in the host gene slyD that absolutely block the lethal and lytic effects of E (14). slyD encodes an FK506 binding protein-type peptidyl-prolyl cis-trans isomerase (PPIase) (15). In vitro, PPIases have been shown to catalyze the folding of proteins limited by peptidyl-prolyl isomerization in their folding pathway. In addition, PPIases have been implicated in protein folding in vivo (16–19). E has five peptidyl-prolyl bonds, including three in the essential hydrophobic domain. The lysis block in the slyD mutant is associated with the failure of E to accumulate in the membrane, suggesting that SlyD is involved in folding or membrane insertion of the lysis protein (T.G.B., unpublished data). E mutants that bypass the slyD lysis defect were isolated by selecting for φX174 plaque formation on a slyD-null lawn. These Epos mutants all were found to contain the missense changes R3H or L19F, or, in the most efficient plaque formers, both changes (W.D.R., unpublished data). Moreover, the Epos protein with both changes accumulates and lyses a slyD host, possibly reflecting the ability of the altered protein to undergo spontaneous folding or membrane insertion, or to access a different folding catalyst. Irrespective of the molecular mechanism by which the pos mutations bypass the SlyD requirement, it seems clear that SlyD cannot be the target of E lytic function. Here we describe the results of a mutant selection undertaken to identify this target.
Materials and Methods
Media, Chemicals, and General Methods.
LB broth was used for all cultures and agar plates (21). When indicated, media was supplemented with ampicillin, chloramphenicol (Cam), kanamycin (Kan), rifampicin, naladixic acid, and tetracycline at final concentrations of 100, 10, 40, 100, 10, and 10 μg/ml, respectively. Isopropyl β-d-thiogalactopyranoside (IPTG) (Alexis, San Diego, CA) was added to a final concentration of 1 mM. Unless otherwise indicated, all chemicals were purchased from Sigma.
DNA Techniques and PCR.
All DNA manipulations were performed according to standard and published procedures (22, 23). F′ DNA was isolated by using the Nucleobond AX 20 Kit (Macherey & Nagel) according to instructions. All enzymes were purchased from Promega with the exception of T4 DNA ligase (Boehringer Mannheim) and Pfu polymerase (Stratagene). Ligation reactions were performed by using the Rapid DNA Ligation Kit from Boehringer Mannheim according to the manufacturer's instructions. Boilates were prepared for chromosomal DNA templates by resuspension of 1 ml of an overnight culture in 100 μl of TE buffer (10 mM Tris⋅HCl, pH 7.6/1 mM EDTA) and boiling for 5 min. The debris was removed by centrifugation before use. PCR compositions unless otherwise indicated are: 10 mM Tris⋅HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, 0.2 mM in each dNTP, 1.5 mM MgCl2, 0.5 μM in each primer, 3 units of Pfu DNA polymerase, and approximately 200 ng of plasmid DNA or 1 μl of a boilate as template. Unless otherwise indicated, cycling parameters were 94°C for 30 sec, 55°C for 30 sec, and 72°C for 2 min for a total of 25 cycles.
Bacterial Strains and Phage.
XL1-Blue (recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 lac [F′∷Tn10 proA+B+ laqIq Δ(lacZ)M15]) was used for all plasmid constructions. As indicated, CCX1 (E. coli C slyD1) (24), Eps4 (CCX1 mraY4), Eps15 (CCX1 mraY15), Eps39 (CCX1 mraY39), RY7281 (CCX1 zac∷Tn10kan), RY7283 (Eps4 zac∷Tn10kan), RY7278 (CCX1 recA1 srl∷Tn10), KL723 (contains F′104 with the 99.8–7.4 min region) (25), CAG12095 (MG1655 zac∷Tn10) (26), CAG12131 (MG1655 zac∷Tn10kan) (26), CQ21 (27), RY7425 (CQ21 zac∷Tn10kan zhd∷Tn10), RY7426 (RY7425 slyD1), RY7427 (RY7425 mraY4), RY7428 (RY7425 slyD1 mraY4), RY7288 (RY7278 rifR), LE392 (hsdR514 supE44 supF58 lacY1 galK2 galT22 metB1 trpR55), RY3270 (LE392 slyD1 zhd∷Tn10 pRY104B), RY3272 (RY3270 mraY4 zac∷Tn10kan), RY7285 (RY7278 [F'104 zac∷Tn10kan mraY4]), RY1275 (Hfr KL16 recA58 srl∷Tn10 ilv val thr spcR [P1tsCam]) (25), and DH5α (φ80 lacZDM15 recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 deoR ΔlacZU169) were used for mutant selection and characterization. φX174Epos4B, referred to as φX174Epos, was isolated as a spontaneous plaque former on a slyD mutant lawn (W.D.R., unpublished results). The Epos4B allele contains both the R3H and L19F missense mutations and henceforth will be referred to as Epos.
Plasmids.
pKN104B is a KanR derivative of pRY104B and contains Epos under control of the tac promoter. pRY104B is identical to pRY100 (24) except that it contains Epos in place of E. pRY104B was digested with ScaI to disrupt bla, and a blunt-ended KanR cassette was inserted. The KanR cassette was obtained from SmaI digestion of pWJC3 and gel purification (28). pEmycZ and pEposmycZ are identical to pRY100 (24) except they contain c-myc-tagged E and Epos, respectively under tac promoter control. pEmycZK is a KanR derivative of pEmycZ constructed in the same manner as pKN104B. pE35GFP is a pQE30 (Qiagen, Chatsworth, CA) derivative containing the E35φgfp fusion under the control of a hybrid lac-T5 phage promoter (W.D.R., unpublished results). pmraY contains mraY under tac control. pmraY4 and pmraY39 are identical to pmraY except they contain the mutant mraY allele indicated by the number. The pmraY series of plasmids was constructed by amplifying the desired mraY allele from boilates of CCX1 and Eps strains with the primers ForMraY (5′-tatcccgggatccccgccatggaagaggtagtacg-3′) and RevMraY (5′-tatcccgggaagcttgccattcatcattcagactgcc-3′). The PCR products were digested with BamHI and HindIII and ligated into appropriately digested pJF118 (29). The pMY30 and pMY304 plasmids are pBAD30 (30) derivatives containing mraY and mraY4, respectively under control of the araBAD promoter. The same PCR products used for pmraY construction were digested with SmaI and HindIII and ligated into appropriately digested pBAD30 (30). pSD1100 is a pACYC184 (31) derivative containing slyD under control of its native promoter. slyD and its upstream promoter region were amplified from the chromosome of MC4100 by PCR using the primers FarForSlyD (5′-cgcggatcctcacgttcgcaaagatgagc-3′) and RevSlyD (5′-gcggatccagggcgagcgcaagcttgaagaaacgccaccgccaca-3′). The PCR product was digested with BamHI and HindIII and ligated into appropriately digested pACYC184.
Selection and Screen for Epos-Resistant Mutants.
A culture of CCX1 pKN104B was grown to an A550 of 0.18, and Epos expression was induced with IPTG. After lysis was complete (approximately 3.5 h), 0.1 ml of the culture was plated on LB-Kan-IPTG to yield approximately 200 colonies per plate. A total of about 2,000 survivors were isolated and screened for φX174Epos phage resistance by using cross-streaks. For cross-streaks, approximately 107 plaque-forming units were spread down the center of a plate and allowed to dry. Survivor colonies were picked directly from the selection plate and streaked across the spread phage. A streak was scored positive if there was significant and reproducible growth across the phage.
General Bacterial and Phage Methods.
Standard bacterial matings were performed essentially as described (21). Triparental matings to generate a merodiploid with eps+ on the chromosome and eps4 on F′104 were performed by mixing 0.5 ml of exponential cultures of KL723 (strain 1), RY7283 (strain 2), and RY7278 (strain 3) and allowing them to stand at 37°C for 5 h. The desired exconjugants were selected by plating dilutions on LB-Kan-tetracycline (Fig. 1). To generate homozygous eps+ merodiploids RY7281 was used as strain 2. P1 transductions were performed essentially as described (21). φX174Epos phage plating was performed as described (24).
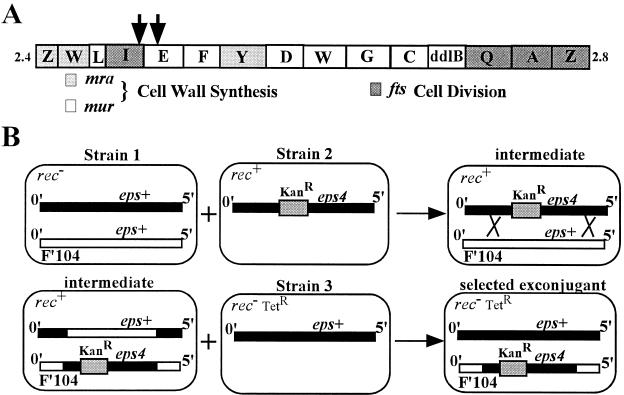
Figure 1.
(A) The mra locus. Shown is a schematic of the mra locus of E. coli present at 2 min. The positions of the mini-Tncam insertions that reduce the ability of F′104eps4 to confer the dominant Eps phenotype are indicated by the arrows. (B) Triparental mating strategy to generate merodiploids. Shown is a schematic of the triparental mating strategy used to generate merodiploids with the eps+ allele on the chromosome and the eps4 allele on F′104. The thick black line represents the 0- to 5-min region of the chromosome. The open line represents the 0- to 5-min region contained on F′104. Relevant alleles present in the 0- to 5-min regions are indicated above the lines. All other relevant genotypic features are indicated in the top left corner of the cartoon cells. The shaded box represents the transposon Tn10kan present at 2 min. The crossed lines represent homologous recombination between the F′ and chromosome. The same strategy was used to generate homozygous eps+ merodiploids by using a strain 2 with eps+ linked to the KanR transposon marker.
Transposon Mutagenesis.
MiniTncam transposon mutagenesis was performed on strain RY7285 (the exconjugant selected from triparental matings) by using the delivery phage λNK1324 essentially as described (32) except the transposition mixture contained 0.5 ml of a 30× concentrated exponential culture of RY7285 in LB-IPTG-10 mM MgSO4. Transposon insertions in the F′ were isolated by mating the pool of transposon mutants with RY2788 and selecting on LB-rifampicin-Kan-Cam. Insertions that eliminated the phage-resistant phenotype conferred by the eps4 allele on the F′ were identified by replica plating for φX174Epos sensitivity. Replica plating was performed by replicating plates containing about 200 colonies on velvet to plates with and without 108 plaque-forming units of φX174Epos.
Inverse PCR.
To identify the location of the transposon insertions that eliminated the dominant phenotype associated with the eps4 allele on F′104, the phage-sensitive exconjugants described above were mated with DH5α and plated on LB-Cam-naladixic acid. The F′104 DNA was purified and digested with RsaI, which cuts twice in the middle of the mini-Tncam transposon and in many locations in the F′. The digested DNA was circularized by ligation. The ligation-dependent and transposon-specific inverse PCR (33) product, containing transposon sequence and sequence immediately outside of the transposon insertion site, was generated by using the primers TnCam1 (5′-gcggatccagggcgagcgctcaggctcaggctctccccgtggagg-3′) and TnCam2 (5′-actggcctcaggcatttgag-3′). Approximately 10 ng of ligated F′ DNA digests was used for the template and cycling parameters were 94°C for 30 sec, 50°C for 30 sec, and 72°C for 3 min for a total of 25 cycles. The product was sequenced by using the primer RevEBam (5′-gcggatccagggcgagcgc-3′). The first 20 bases of good sequence outside of the transposon were used to search the E. coli genome by using the blastn program (www.ncbi.nlm.nih.gov).
DNA Sequencing.
Sequencing PCRs contained approximately 50 ng of PCR product, 1.4 μM sequencing primer, 1.3 μl of 5× reaction buffer (400 mM Tris/10 mM MgCl2), and 0.7 μl of Applied Biosystems Big-Dye reaction mixture in a total reaction volume of 7 μl. Sequencing was otherwise identical to published methods (23). The entire mraY genes of CCX1 and Eps39 were sequenced from PCR products generated from their chromosomal DNA. The PCR products were sequenced by using the primers ForMraY, RevMraY, MYForIN (5′-agttactcttcatttgtcgccggtggttttgcg-3′), MYRevIN (5′-agttactcttcacacaatgacgcgcggttccgg-3′), MYForOut (5′-agttactcttcagtgcgtttctggattatttcg-3′), and MYRevOut (5′-agttactcttcaaaatacggtcggcataattgc-3′). This procedure gave good sequence for both strands of the entire gene, and the deletion of codon 172 was the only mutation observed in mraY39. For mraY4 and mraY15 the sequence of both strands of an approximately 400-bp fragment surrounding the mutation was determined and found to contain the T to C transition in codon 288. This fragment, generated by PCR from the mutant chromosomes using the primers MYForIN and MYRevIN, conferred the φX174 Epos phage plating phenotype to CCX1 when it replaced the wild-type (wt) fragment of pmraY. This cassette exchange was performed by using the Seamless Cloning Kit (Stratagene) and the primers MYForOut, MYRevOut, MYForIN, and MYRevIN according to the instructions. The fragment was identified because the mutations in mraY4 and mraY15 fortuitously destroy an EcoRI site contained within it.
Lysis Profiles.
Lysis profiles were generated as described (24). Briefly, cultures were grown in LB-ampicillin or LB-Kan-ampicillin at 37°C to an A550 of 0.2 to 0.4 and induced with IPTG for expression of the E alleles. A550 measurements were continued until lysis was essentially complete.
Membrane Topology Prediction.
The membrane topology of MraY was predicted by using a hidden Markov model (34) at the tmhmm server (www.cbs.dtu.dk/services/TMHMM-1.0/).
Results and Discussion
Selection for Epos-Resistant Mutants.
To identify the gene encoding the target of E lysis we selected for spontaneous mutants of CCX1, E. coli C slyD1, that gain resistance to Epos expression from the plasmid pKN104B. The majority of the selected survivors contained plasmid mutations that eliminated Epos expression. To identify host mutants conferring resistance to Epos in the high background of plasmid mutants, we screened the survivors by cross-streaking them against the φX174Epos phage. Survivors resulting from plasmid mutations are still sensitive to lysis by the phage-encoded Epos and are thus phage sensitive. We expected true Epos-resistant host mutants to be resistant to Epos from the phage as well as the plasmid. Approximately 2,000 survivors were screened, and 17 eps (Epos sensitivity) mutants scored positive for phage resistance. Two types of eps mutants were isolated, 14 with a partial phage-resistance phenotype and three with a tight resistance phenotype. We focused on the three tight mutant strains, Eps4, Eps15, and Eps39, for this investigation. Except for Eps39, the tight mutant strains exhibited no growth or colony morphology phenotypes other than lysis resistance. Eps39 appeared to be somewhat unstable, yielding both small and large colonies upon restreaking, which may indicate the accumulation of suppressor mutations. However, both types of colonies remained resistant to the phage in cross-streaks.
Phenotypic Characterization of the eps Mutants.
Each of the eps mutants were completely resistant to Epos expressed from pKN104B (data not shown). To ensure that the pKN104B plasmids from the mutant strains were still functional, their plasmid DNA was isolated and transformed into the parental strain CCX1. All plasmids lysed CCX1 with normal efficiency (data not shown), indicating that the mutants did not survive the selection as a result of a plasmid mutation. The pKN104B plasmid was cured from the mutant strains and the effect of the mutations on the plating efficiency of φX174Epos was determined (Table 1). As shown, the plating efficiency of φX174Epos was significantly reduced for the eps mutants compared with the parental CCX1 strain. In addition, the average plaque diameter was significantly smaller on eps mutant lawns. A one-step growth experiment was performed, and φX174Epos phage accumulation was observed in the Eps4 mutant strain (data not shown). This indicates that, at least in the case of the eps4 allele, the mutation affects only lysis and not phage development. Introduction of pSD1100, a medium copy plasmid containing slyD under control of its chromosomal promoter, suppressed the φX174Epos plating defect of the mutants (data not shown; see below).
Table 1.
φX174 Epos efficiency of plating (e.o.p.) on mutant strains
| Strain | e.o.p. | Plaque size, mm |
|---|---|---|
| CCX1 | 1 | 3–4 |
| Eps4 | 0.08 | Pin-point |
| Eps15 | 0.09 | Pin-point |
| Eps39 | 0.3 | 0.5–1 |
| CCX1 pJF118 | 1 | 3–4 |
| CCX1 pmraY+ | 0.8 | 2–3 |
| CCX1 pmraY4 | 0.04 | 0.5–1 |
| CCX1 pmraY39 | 0.06 | 0.5–1 |
Genetic Mapping of the eps Mutations.
Hfr and P1 mapping localized the eps mutations to the 2-min region of the E. coli chromosome (60% cotransducible with a Tn10 marker at 2 min). The 2-min region contains the mra locus that is rich in genes for cell wall synthesis and cell division (Fig. 1A) (35, 36). To assess the recessive or dominance of the eps4 allele, a triparental mating of F′104 was used to generate merodiploids of the 0- to 5-min region of the chromosome (Fig. 1B). Merodiploids containing two wt copies of the 0- to 5-min region showed normal φX174 Epos plating efficiency and plaque size. However, merodiploids containing an eps+ allele on the chromosome and an eps4 allele on F′104 had the phage-resistant phenotype. Therefore, the eps4 allele is dominant over wt.
The eps Mutations Are Located in mraY.
To identify the gene containing the dominant eps4 mutation, a strain carrying F′104eps4 was mutagenized with mini-Tncam and mated with an eps+ strain. Exconjugants, carrying transposon insertions in the F′, were screened for φX174Epos sensitivity by replica plating. The positions of mini-Tncam insertions that eliminated the phage-resistance phenotype associated with the F′ were determined by inverse PCR and sequencing. Two insertions mapping to ftsI and murE were obtained (Fig. 1A), both with partial phage sensitivity, suggesting the insertions were polar on the eps locus rather than knockouts. Knowing that the lytic function of E is contained in its hydrophobic membrane domain, we reasoned that its cellular target should be a membrane protein. The first gene downstream of the transposon insertions encoding a membrane protein is mraY. We amplified the mraY alleles from the parental and mutant strains and inserted them under control of the tac promoter in the vector pJF118 (29). As shown in Table 1, basal expression of mraY cloned from the mutant strains (pmraY4 and pmraY39) conferred the φX174Epos plating defect to the parental strain CCX1. On the other hand, basal expression of mraY cloned from the parental strain (pmraY+) had only a slight phage-plating defect. Therefore, dominant mutations in mraY, a gene encoding a membrane-bound enzyme involved in cell wall synthesis, confer the Eps phenotype. We therefore have renamed the eps alleles as mraY4, mraY15, and mraY39. Interestingly, it appears that basal expression of mraY39 from the plasmid in conjunction with mraY+ from the chromosome confers a more severe phage-plating defect than the mraY39 allele alone in the chromosome (compare the efficiency of plating for Eps39 with that of CCX1 pmraY39). We do not currently have an explanation for this observation.
The Lysis Phenotype of the mraY4 Allele Is General.
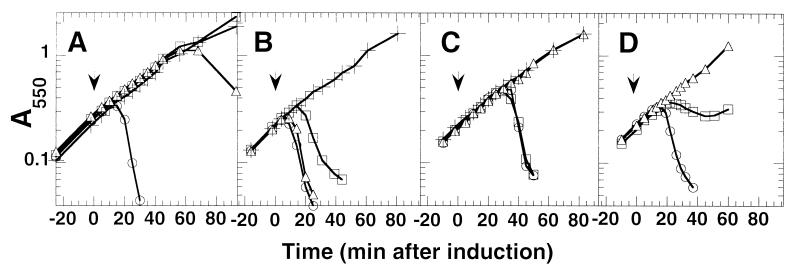
The dominant mraY mutants were isolated by selecting for Epos resistance in an E. coli C slyD1 background. We therefore were interested in determining whether the lysis-defective phenotype of the mraY4 allele is general and whether it depended on the allelic state of E and slyD. The mraY4 allele was transduced into E. coli K-12 slyD+ and slyD1 strains, and the lysis phenotype was assessed by inducing expression of various plasmid-borne E alleles. The mraY4 allele severely delayed the lysis of wt E in a slyD+ background and did not affect the E resistance of the slyD1 strain (Fig. 2A). Interestingly, only the slyD1 mraY4 double mutant was resistant to Epos (Fig. 2B). This finding is consistent with the observation that slyD, in multicopy, suppresses the φX174 Epos plating defect of the original mutants (see above). Epos is more lytic than the wt E allele in a slyD+ strain (compare lysis in Fig. 2 A and B). Apparently, Epos is so efficient in a slyD+ strain that the mraY4 allele has no effect on its ability to cause lysis. However, when the efficiency of Epos is reduced because of a slyD1 null mutation, the presence of the mraY4 allele completely inhibits lysis (Fig. 2B). The mraY4 allele abolishes lysis by E35φgfp irrespective of the allelic state of slyD (Fig. 2C). Unlike E, lytic chimera, in which the C-terminal two-thirds of the E protein is replaced by a variety of protein domains, are insensitive to the allelic state of slyD (W.D.R., unpublished data; ref. 14). This finding has led to criticism that the E fusions are acting by a mechanism distinct from E (37). However, the mraY4 allele affects the lysis of the E35φgfp fusion in addition to E and Epos, arguing that all of the E alleles, with or without the fusion domains, are acting via the same mechanism and differ only with respect to their slyD dependence.
Figure 2.
The mraY4 allele causes a general lysis defect. (A) Lysis profiles from pEmycZ induction in different genetic backgrounds. The allelic state of slyD and mraY in each strain is as follows: slyD+ mraY+ (RY7425) (○), slyD1 mraY+ (RY7426) (□), slyD+ mraY4 (RY7427) (▵), and slyD1 mraY4 (RY7428) (+). The arrow indicates the time of IPTG addition to induce lysis gene expression. (B) The strains are the same as indicated in A except they contain the plasmid pEposmycZ. (C) Same as in A except strains contain the plasmid pE35GFP. (D) Lysis profiles resulting from increased mraY expression. Emyc expression was induced from CAG12095 pBAD30 pEmycZK (○), pMY30 pEmycZK (□), and pMY304 pEmycZK (▵) at time 0. All cultures were grown in the presence of 0.2% arabinose for constitutive expression of the mraY alleles cloned in the pBAD30 vector.
Sequence of the mraY Mutations.
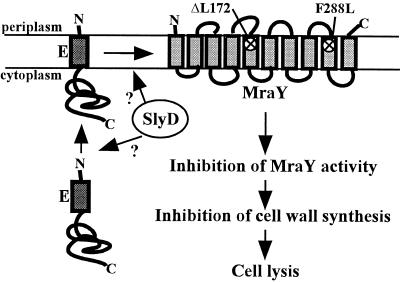
mraY is an essential E. coli gene (38). It encodes a 360-aa membrane-bound enzyme, often referred to as translocase I, that catalyzes the formation of the first lipid-linked cell wall precursor, undecaprenol-pyrophospho-N-acetylmuramic acid-pentapeptide or lipid I (39). MraY belongs to a ubiquitous superfamily of membrane-bound enzymes that catalyze the transfer of an aminosugar-phosphate from UMP in the cytoplasm to polyisoprenyl-phosphate in the membrane (40). The enzymology of MraY is well established but little is known about its molecular structure (41). MraY has 10 stretches of hydrophobic amino acids predicted to be TMDs and a predicted topology that is supported by recent bla fusion analysis (Fig. 3) (42). This topology is also consistent with the topology determined for a hamster enzyme of the same superfamily for which there is also evidence suggesting oligomeric structure (43, 44). Sequencing analysis revealed that both the mraY4 and mraY15 alleles have a change creating a F288L substitution near the boundary of predicted TMD 9. The mraY39 allele was found to have an in-frame deletion of codon 172, located near the boundary of predicted TMD 5 (Fig. 3). Given that the hydrophobic N terminus of the E protein is the lytic domain, it is not surprising that both mraY mutations conferring lysis resistance cause changes in the amino acid sequence of predicted TMDs in MraY. Also, considering that the mraY mutants are viable, the mutations must not seriously affect the essential enzymatic function of MraY.
Figure 3.
Proposed model for the mechanism of E-mediated cell lysis. The E protein is synthesized in the cytoplasm and integrated into the inner membrane. The cytoplasmic SlyD PPIase is required for E protein accumulation, either before or after E reaches the membrane. We propose that as E accumulates in the membrane it interacts with and inhibits MraY. Inhibition of MraY activity would result in the inhibition of cell wall synthesis and cell lysis. The predicted topology of MraY in the membrane is drawn with the positions of the mutations affecting lysis indicated with an x. The topology of the E protein is drawn according to the observation that the C terminus is located in the cytoplasm (R.Y., unpublished data), although it is not known whether the N terminus of E actually traverses the membrane.
A Model for the Mechanism of E-Mediated Lysis.
The isolation of lysis-resistance mutations in mraY strongly suggests that E-mediated lysis is caused by an inhibition or derangement of cell wall synthesis. The depletion of MraY from cells causes lysis, as does the inhibition of MraY by the antibiotic mureidomycin (38, 45). Taken together, these data suggest that the target of the E lysis protein is MraY itself (Fig. 3). However, we cannot discount the alternative model that E complexes with undecaprenol-phosphate, making it unavailable as a substrate for MraY. In both cases MraY activity would be depleted from the cell. The simplest idea, that E titrates out the activity of MraY by binding as an inhibitor, leads to the prediction that increasing the cellular MraY concentration would delay or even inhibit lysis. To test this prediction, mraY was cloned into a medium copy plasmid under control of the araBAD promoter. Expression of mraY from this plasmid in conjunction with E from the tac promoter on a compatible plasmid, severely delayed lysis (Fig. 2D). Expression of mraY4 from the same plasmid completely inhibited lysis (Fig. 2D). Thus, as predicted, increasing the level of MraY causes a significant defect in E lysis. The dominance of the mraY alleles is also consistent with MraY inhibition. A mutant mraY producing a protein that cannot be inhibited by E would be expected to confer dominant lysis resistance over wt mraY, producing a protein that is sensitive to E inhibition.
The most likely mode by which E or E fusions could inhibit MraY would be through a direct interaction mediated by contacts between the hydrophobic domain of E and one or more of the TMDs of the oligotopic enzyme. This would explain why the lysis-resistance changes in MraY are localized to putative TMDs. These changes may abolish favorable contacts between E and MraY and alter the interaction of the two proteins.
From the perspective of this model, the failure of E lysis in the slyD1 background and the failure of the mraY mutations to block E or Epos lysis in the slyD+ background are related phenotypes pointing to the role of the SlyD PPIase in lysis protein accumulation. In a slyD host, E fails to accumulate at all, leaving MraY activity unaffected, whereas in a slyD+ host, the presence of active SlyD allows so much E or Epos to accumulate that the reduced sensitivity conferred by the eps changes in MraY are overwhelmed and its activity is inhibited (Fig. 3). SlyD could be promoting lysis protein accumulation in a number of ways. It may be directly involved in the folding, stabilization, or membrane insertion of E. Alternatively, E may be unstable unless it is complexed with MraY, and SlyD may promote this interaction, leading to an indirect stabilization of E.
Other Models for E Function.
The results reported here are completely inconsistent with the “transmembrane tunnel” model (refs. 13, 37, 46, and 47 and references therein) for E-mediated lysis. In this model, it is proposed that the tunnel, which spans the entire envelope, consists of an oligomer of E protein. Formation of the tunnel is proposed to cause a rapid release of cellular osmotic pressure, resulting in expulsion of the cytosolic contents, including progeny virions (13). A role for SlyD also has been proposed, in which the PPIase activity is involved in catalyzing a dramatic conformational change that leaves the N terminus of E embedded in the inner membrane and externalizes the C-terminal domain from the cytoplasm so it can span the rest of the envelope (46). This makes no sense in view of the isolation of lysis-resistant mutants of mraY. The simple primary structure of E and the degeneracy of its C-terminal requirements make it highly unlikely that it could form aqueous pores of such unprecedented size. Instead, we believe the large holes reported by Witte et al. (13) reflect the consequence of E-mediated inhibition of murein synthesis, which probably manifests itself as weak areas in the envelope. The prevalence of septal locales for these lesions and the established dependence of E-mediated lysis on continued cellular septation probably reflect the fact that the developing septum consists entirely of newly synthesized cell wall (48) and is thus highly susceptible to an interruption of the supply of peptidoglycan precursors. In contrast, during cellular elongation, new murein synthesis occurs at disperse sites in the pre-existing cell wall (5, 48). A lack of cell wall precursors at these sites is thus less likely to be rapidly catastrophic.
Is Cell Wall Synthesis Targeted by All Simple Lytic Phages?
The idea that the single lysis gene of the simple lytic phage φX174 acts as a “protein antibiotic” not unlike other cell wall synthesis inhibitors suggests that there may be a unifying principle in the two fundamental modes by which bacteriophage achieve host lysis and dispersal of progeny. That is, it is fundamentally necessary to subvert the continuity of the cell wall. This can be done either by elaborating a multigene holin-endolysin system to achieve properly timed degradation of the cell wall (1), or by poisoning cell wall synthesis during cell growth. There are two other known single-gene lysis systems, the L gene of group I and II RNA phages and the bifunctional A2 maturation-lysis gene of the group III and IV RNA phages (20). Neither of these lysis genes are associated with active cell wall degradation. It will be of interest to see whether these single-gene lysis systems also act by attacking the cell wall synthesis machinery, and if so, at which step.
Acknowledgments
We thank all of the members of the Young laboratory, past and present, for their help and encouragement. Special thanks to Debby Siegele and Kevin Young for strains and technical assistance. The patient colony streaking of Thuyen Nguyen and April Stanley is gratefully acknowledged. This work was supported by Public Health Service Grant GM27099 and funds from the Robert A. Welch Foundation and the Texas Agricultural Experiment Station.
Abbreviations
- PPIase
peptidyl-prolyl cis-trans isomersase
- Cam
chloramphenicol
- Kan
kanamycin
- IPTG
isopropyl β-d-thiogalactopyranoside
- TMD
transmembrane domain
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Young R. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanger F, Air G M, Barrell B G, Brown N L, Coulson A R, Fiddes J C, Hutchison C A, III, Slocombe P M, Smith M. Nature (London) 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 3.Young K D, Young R. J Virol. 1982;44:993–1002. doi: 10.1128/jvi.44.3.993-1002.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrich B, Lubitz W, Plapp R. Mol Gen Genet. 1982;185:493–497. doi: 10.1007/BF00334146. [DOI] [PubMed] [Google Scholar]
- 5.Burman L G, Raichler J, Park J T. J Bacteriol. 1983;155:983–988. doi: 10.1128/jb.155.3.983-988.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckley K J, Hayashi M. Mol Gen Genet. 1986;204:120–125. doi: 10.1007/BF00330198. [DOI] [PubMed] [Google Scholar]
- 7.Bläsi U, Lubitz W. J Gen Virol. 1985;66:1209–1213. doi: 10.1099/0022-1317-66-6-1209. [DOI] [PubMed] [Google Scholar]
- 8.Bradley D E, Dewar C A, Robertson D. J Gen Virol. 1969;5:113–121. doi: 10.1099/0022-1317-5-1-113. [DOI] [PubMed] [Google Scholar]
- 9.Bläsi U, Halfmann G, Lubitz W. In: Microbial Cell Wall Synthesis and Autolysis. Nombela C, editor. Amsterdam: Elsevier; 1984. pp. 213–218. [Google Scholar]
- 10.Hahn F E, Ciak J. Science. 1957;125:119–120. doi: 10.1126/science.125.3238.119. [DOI] [PubMed] [Google Scholar]
- 11.Roof W D, Young R. J Bacteriol. 1993;175:3909–3912. doi: 10.1128/jb.175.12.3909-3912.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubitz W, Harkness R E, Ishiguro E E. J Bacteriol. 1984;159:385–387. doi: 10.1128/jb.159.1.385-387.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witte A, Wanner G, Bläsi U, Halfmann G, Szostak M, Lubitz W. J Bacteriol. 1990;172:4109–4114. doi: 10.1128/jb.172.7.4109-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maratea D, Young K, Young R. Gene. 1985;40:39–46. doi: 10.1016/0378-1119(85)90022-8. [DOI] [PubMed] [Google Scholar]
- 15.Roof W D, Horne S M, Young K D, Young R. J Biol Chem. 1994;269:2902–2910. [PubMed] [Google Scholar]
- 16.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Nature (London) 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 17.Dartigalongue C, Raina S. EMBO J. 1998;17:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Missiakas D, Betton J-M, Raina S. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 19.Danese P N, Silhavy T J. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 20.van Duin J. In: The Bacteriophages. Calendar R, editor. New York: Plenum; 1988. pp. 117–167. [Google Scholar]
- 21.Miller J H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 22.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 23.Smith D L, Chang C-Y, Young R. Gene Exp. 1998;7:39–52. [PMC free article] [PubMed] [Google Scholar]
- 24.Roof W D, Fang H Q, Young K D, Sun J, Young R. Mol Microbiol. 1997;25:1031–1046. doi: 10.1046/j.1365-2958.1997.5201884.x. [DOI] [PubMed] [Google Scholar]
- 25.Low K B. Bacteriol Rev. 1972;36:587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks J, Grossman A D, Erichson J W, Gross C A. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang Y F, Ma D P, Young R, Struck D K. DNA Seq. 1993;3:357–367. doi: 10.3109/10425179309020837. [DOI] [PubMed] [Google Scholar]
- 28.Chen W-J, Gross L, Joho K E, McAllister W T. Gene. 1992;111:143–144. doi: 10.1016/0378-1119(92)90617-x. [DOI] [PubMed] [Google Scholar]
- 29.Fürste J P, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 30.Guzman L-M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang A C Y, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleckner N, Bender J, Gottesman S. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 33.Ochman H, Gerber A S, Hartl D L. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonnhammer E L, von Heijne G, Krogh A. Proc Sixth Int Conf Intelligent Systems Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 35.Hara H, Yasuda S, Horiuchi K, Park J T. J Bacteriol. 1997;179:5802–5811. doi: 10.1128/jb.179.18.5802-5811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mengin-Lecreulx D, Ayala J, Bouhss A, van Heijenoort J, Parquet C, Hara H. J Bacteriol. 1998;180:4406–4412. doi: 10.1128/jb.180.17.4406-4412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witte A, Bläsi U, Halfmann G, Szostak M, Wanner G, Lubitz W. Biochimie. 1990;72:191–200. doi: 10.1016/0300-9084(90)90145-7. [DOI] [PubMed] [Google Scholar]
- 38.Boyle D, Donachie W. J Bacteriol. 1998;180:6429–6432. doi: 10.1128/jb.180.23.6429-6432.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikeda M, Wachi M, Jung H, Ishino F, Matsuhashi M. J Bacteriol. 1991;173:1021–1026. doi: 10.1128/jb.173.3.1021-1026.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehrman M A. Glycobiology. 1994;4:768–771. doi: 10.1093/glycob/4.6.768. [DOI] [PubMed] [Google Scholar]
- 41.Brandish P, Burnham M, Lonsdale J, Southgate R, Inukai M, Bugg T. J Biol Chem. 1996;271:7609–7614. doi: 10.1074/jbc.271.13.7609. [DOI] [PubMed] [Google Scholar]
- 42.Bouhss A, Mengin-Lecreulx D, Le Beller D, van Heijenoort J. Mol Microbiol. 1999;34:576–585. doi: 10.1046/j.1365-2958.1999.01623.x. [DOI] [PubMed] [Google Scholar]
- 43.Dan N, Lehrman M A. J Biol Chem. 1997;272:14214–14219. doi: 10.1074/jbc.272.22.14214. [DOI] [PubMed] [Google Scholar]
- 44.Dan N, Middleton R B, Lehrman M A. J Biol Chem. 1996;271:30717–30724. doi: 10.1074/jbc.271.48.30717. [DOI] [PubMed] [Google Scholar]
- 45.Isono F, Inukai M, Takahashi S, Haneishi T, Kinoshita T, Kuwano T. J Antibiot. 1989;42:674–679. doi: 10.7164/antibiotics.42.667. [DOI] [PubMed] [Google Scholar]
- 46.Witte A, Schrot G, Schon P, Lubitz W. Mol Microbiol. 1997;26:337–346. doi: 10.1046/j.1365-2958.1997.5781941.x. [DOI] [PubMed] [Google Scholar]
- 47.Witte A, Brand E, Mayrhofer P, Narandja F, Lubitz W. Arch Microbiol. 1998;170:259–268. doi: 10.1007/s002030050641. [DOI] [PubMed] [Google Scholar]
- 48.de Pedro M A, Quintela J C, Holtje J V, Schwarz H. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]