Abstract
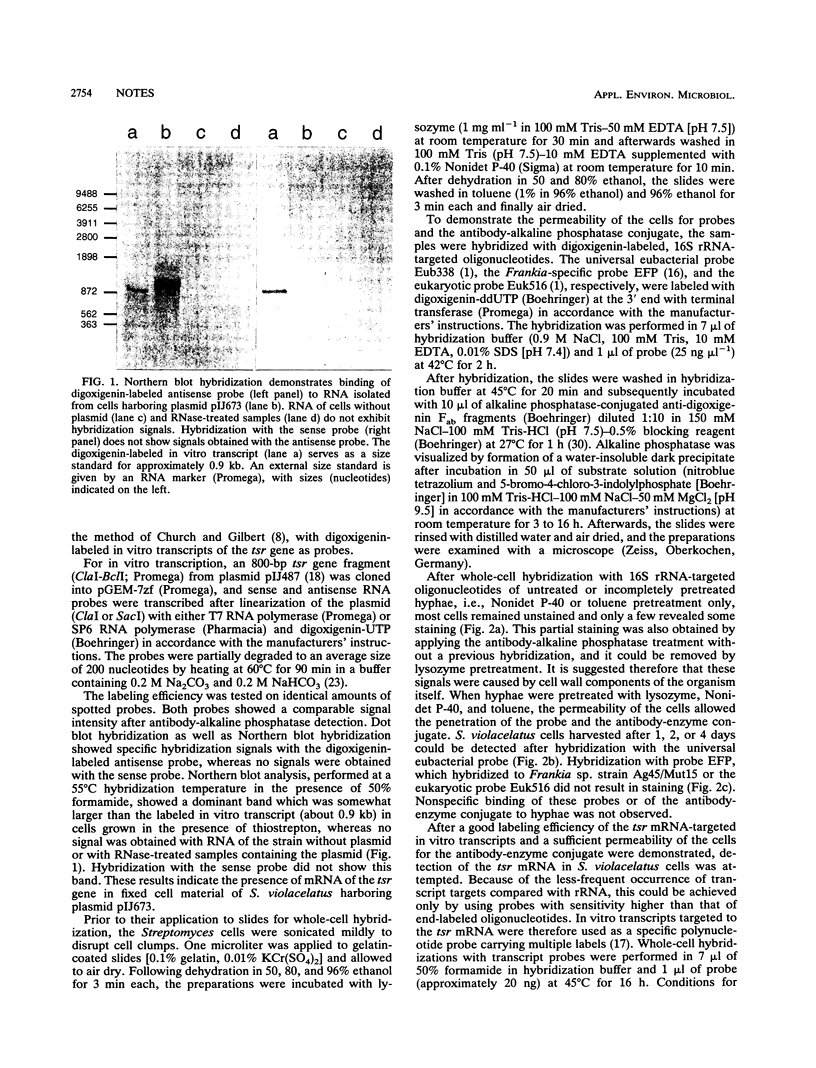
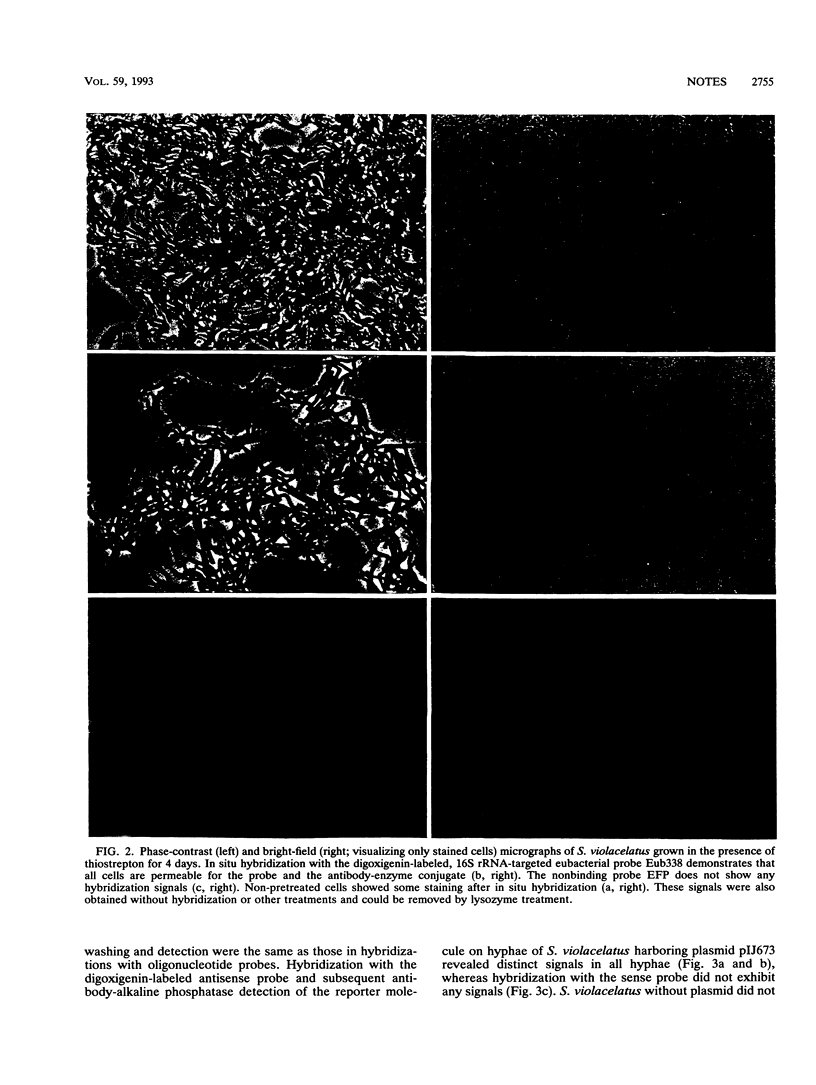
Detection of mRNA of the thiostrepton resistance gene (tsr) harbored by plasmid pIJ673 in Streptomyces violacelatus was achieved by whole-cell hybridization with digoxigenin-labeled in vitro transcripts followed by an antibody-alkaline phosphatase detection of the digoxigenin reporter molecule. Prior to hybridization, the cells had to be permeabilized by lysozyme, the detergent Nonidet P-40, and toluene. The permeability of the S. violacelatus cells for probes and the antibody-alkaline phosphatase conjugate was demonstrated by hybridization with digoxigenin-labeled, 16S rRNA-targeted oligonucleotides.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990 Jun;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Stromley J., Devereux R., Key R., Stahl D. A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992 Feb;58(2):614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Baumgartner M., Dutrillaux B., Lemieux N., Lilienbaum A., Paulin D., Viegas-Péquignot E. Genes occupy a fixed and symmetrical position on sister chromatids. Cell. 1991 Feb 22;64(4):761–766. doi: 10.1016/0092-8674(91)90505-s. [DOI] [PubMed] [Google Scholar]
- Celeda D., Bettag U., Cremer C. PCR amplification and simultaneous digoxigenin incorporation of long DNA probes for fluorescence in situ hybridization. Biotechniques. 1992 Jan;12(1):98–102. [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundliffe E. How antibiotic-producing organisms avoid suicide. Annu Rev Microbiol. 1989;43:207–233. doi: 10.1146/annurev.mi.43.100189.001231. [DOI] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Distel D. L., DeLong E. F., Waterbury J. B. Phylogenetic characterization and in situ localization of the bacterial symbiont of shipworms (Teredinidae: Bivalvia) by using 16S rRNA sequence analysis and oligodeoxynucleotide probe hybridization. Appl Environ Microbiol. 1991 Aug;57(8):2376–2382. doi: 10.1128/aem.57.8.2376-2382.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D., Amann R. I., Ludwig W., Akkermans A. D., Schleifer K. H. Detection of micro-organisms in soil after in situ hybridization with rRNA-targeted, fluorescently labelled oligonucleotides. J Gen Microbiol. 1992 May;138(5):879–887. doi: 10.1099/00221287-138-5-879. [DOI] [PubMed] [Google Scholar]
- Höltke H. J., Kessler C. Non-radioactive labeling of RNA transcripts in vitro with the hapten digoxigenin (DIG); hybridization and ELISA-based detection. Nucleic Acids Res. 1990 Oct 11;18(19):5843–5851. doi: 10.1093/nar/18.19.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T., Hopwood D. A., Wright H. M., Thompson C. J. pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol Gen Genet. 1982;185(2):223–228. doi: 10.1007/BF00330791. [DOI] [PubMed] [Google Scholar]
- Scheres B., Van De Wiel C., Zalensky A., Horvath B., Spaink H., Van Eck H., Zwartkruis F., Wolters A. M., Gloudemans T., Van Kammen A. The ENOD12 gene product is involved in the infection process during the pea-Rhizobium interaction. Cell. 1990 Jan 26;60(2):281–294. doi: 10.1016/0092-8674(90)90743-x. [DOI] [PubMed] [Google Scholar]
- Scheres B., van Engelen F., van der Knaap E., van de Wiel C., van Kammen A., Bisseling T. Sequential induction of nodulin gene expression in the developing pea nodule. Plant Cell. 1990 Aug;2(8):687–700. doi: 10.1105/tpc.2.8.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Köhler M., Vogt P., von Malsch K., Schweizer D. Chromosomal in situ hybridization with double-labeled DNA: signal amplification at the probe level. Cytogenet Cell Genet. 1992;60(1):4–7. doi: 10.1159/000133282. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Skinner R. H., Thompson J., Ward J. M., Hopwood D. A., Cundliffe E. Biochemical characterization of resistance determinants cloned from antibiotic-producing streptomycetes. J Bacteriol. 1982 Aug;151(2):678–685. doi: 10.1128/jb.151.2.678-685.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultsch A., Schuster C. M., Betz H., Wisden W. In situ hybridization with oligonucleotides: a simplified method to detect Drosophila transcripts. Nucleic Acids Res. 1991 Jul 11;19(13):3746–3746. doi: 10.1093/nar/19.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Wiel C., Norris J. H., Bochenek B., Dickstein R., Bisseling T., Hirsch A. M. Nodulin Gene Expression and ENOD2 Localization in Effective, Nitrogen-Fixing and Ineffective, Bacteria-Free Nodules of Alfalfa. Plant Cell. 1990 Oct;2(10):1009–1017. doi: 10.1105/tpc.2.10.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarda B., Amann R., Wallner G., Schleifer K. H. Identification of single bacterial cells using digoxigenin-labelled, rRNA-targeted oligonucleotides. J Gen Microbiol. 1991 Dec;137(12):2823–2830. doi: 10.1099/00221287-137-12-2823. [DOI] [PubMed] [Google Scholar]






