Abstract
The newly described heterotrophic estuarine dinoflagellate Pfiesteria piscicida has been linked with fish kills in field and laboratory settings, and with a novel clinical syndrome of impaired cognition and memory disturbance among humans after presumptive toxin exposure. As a result, there is a pressing need to better characterize the organism and these associations. Advances in Pfiesteria research have been hampered, however, by the absence of genomic sequence data. We employed a sequencing strategy directed by heteroduplex mobility assay to detect Pfiesteria piscicida 18S rDNA “signature” sequences in complex pools of DNA and used those data as the basis for determination of the complete P. piscicida 18S rDNA sequence. Specific PCR assays for P. piscicida and other estuarine heterotrophic dinoflagellates were developed, permitting their detection in algal cultures and in estuarine water samples collected during fish kill and fish lesion events. These tools should enhance efforts to characterize these organisms and their ecological relationships. Heteroduplex mobility assay-directed sequence discovery is broadly applicable, and may be adapted for the detection of genomic sequence data of other novel or nonculturable organisms in complex assemblages.
Keywords: harmful algal blooms, Pfiesteria shumwayae
Harmful algal blooms with adverse environmental, economic, and human health effects have been recognized with increasing frequency (1–4). For example, the heterotrophic dinoflagellate Pfiesteria piscicida has been repeatedly linked to fish kills in North Carolina and Chesapeake Bay estuarine waters (5–8). Association between exposure to Pfiesteria and human health effects, including impaired concentration and learning, has brought this organism into the public health spotlight as well (8–10). These observations initiated broad new efforts to characterize the organism, its associated toxicity, and its impact on human health.
Rapid identification of P. piscicida in environmental samples and dinoflagellate cultures has been problematic. The organism has a complex life cycle, and its presence in the water column is sometimes ephemeral (5–8, 11–13). The new species P. piscicida was described based on scanning electron microscope images of its distinctive thecal plate arrangement (6), but at the level of resolution of light microscopy the organism lacks features that distinguish it from a number of other small heterotrophic dinoflagellates. Thus, “pfiesteria-like organisms” present in water samples can be counted by using light microscopy, but definitive identification of P. piscicida has required scanning electron microscopy (SEM) and thecal plate analysis. This labor-intensive approach requires culture amplification using either algal or fish prey, and, in this context, undesired growth of competing algal strains or species may occur. Development of molecular techniques for identification of P. piscicida and related species has, consequently, been hampered. To address these problems, we adopted a sequence discovery approach designed to permit detection of dinoflagellate DNA “signatures” consistently associated with phenotypes of interest (for instance, icthyotoxicity in dinoflagellate/fish bioassays). We selected ribosomal (18S) rDNA gene sequences as the target for amplification and characterization because of their wide use in the phylogenetic evaluation of microorganisms, including dinoflagellates (14–19). Highly conserved sequences within dinoflagellate SSU genes permitted design of a “phylum-selective” PCR primer pair with high selectivity for dinoflagellates. Amplified 18S rDNA sequences thus enriched for dinoflagellate DNA were derived from both dinoflagellate cultures and environmental samples (i.e., DNA extracted from filtered estuarine water samples). These PCR products were then assessed by heteroduplex mobility assay (HMA). The assay is based on the property of DNA heteroduplexes to migrate more slowly through polyacrylamide gels than homoduplexes. HMA can be used to detect single nucleotide differences between DNA fragments in human genetic counseling (20). In the work described herein, the assay readily detected sequence diversity among dinoflagellate 18S PCR amplicon pools. Use of this assay permitted determination of the P. piscicida full-length 18S rDNA sequence, and development of species-specific primers and probes for a PCR based detection system.
Use of sequence based methods for identification of heterotrophic estuarine dinoflagellates may help resolve current ambiguities related to these organisms and their impact on estuarine systems and human health. For instance, a variety of dinoflagellate species found in mid-Atlantic estuarine waters, including P. piscicida, a second “pfiesteria-like” dinoflagellate described herein as “Pfiesteria species B” (the name Pfiesteria shumwayae has been proposed in a recently submitted formal naming paper), Gyrodinium galatheanum (21), and the parasitic dinoflagellate Amyloodinium ocellatum (22) may all be associated with fish kill events or fish disease. Other species likely contribute to such events as well. Development of molecular approaches that permit rapid identification of dinoflagellates of interest complemented by sequence discovery methods for identification of novel species should enhance efforts to characterize these organisms and their environmental impact.
Materials and Methods
Dinoflagellate Cultures.
Cultures of P. piscicida and pfiesteria-like dinoflagellates were established from samples collected in estuarine waters along the Atlantic coast from Florida to Maryland (Table 1; also see Table 2, published as supplemental data on the PNAS web site, www.pnas.org). Clonal cultures were established through microtransfer techniques and were maintained by using previously described methods (5–7). The pfiesteria-like dinoflagellates were identified as such by light microscopy. Species identification was by plate tabulation, under scanning electron microscopy (SEM). Fish kill (toxicity) bioassays were performed with some cultures as indicated (Table 1) by using previously described methods (6, 8, 13). Cultures were obtained from the Aquatic Botany Laboratory of North Carolina State University (NCSU), the Provasoli-Guillard National Center for Culture of Marine Phytoplankton (CCMP), and the Florida Department of Environmental Protection. Additional dinoflagellate reference cultures (from CCMP, n = 33) and uncharacterized pfiesteria-like dinoflagellate cultures (from CCMP, n = 25) were obtained for performance testing of the PCR assays described herein (Table 2 in the supplemental data).
Table 1.
Dinoflagellate cultures characterized by fish bioassay, microscopy, and selective PCR
| Sample | Characterization* | Fish bioassay | P. piscicida PCR | Species B PCR |
|---|---|---|---|---|
| NCSU:Chic 97-1 | P. piscicida (S) | + | + | − |
| NCSU:113-2 | P. piscicida (S) | + | + | − |
| NCSU:114-1-5 | P. piscicida (S) | + | + | − |
| NCSU:125-4 | P. piscicida (S) | + | + | − |
| NCSU:102-1 | P. piscicida (S) | + | + | − |
| NCSU:97-1 | P. piscicida (S) | + | + | − |
| Florida DEP† | P. piscicida (S) | − | + | − |
| NCSU:98-3-B | Pfiesteria-like (L) | − | − | − |
| NCSU:7-28-T | Pfiesteria species B (S) | + | − | + |
| NCSU:15-T | Pfiesteria species B (S) | + | + | + |
| NCSU:BP | Pfiesteria species B (S) | + | − | + |
| NCSU:142 | Cryptoperidiniopsis (S) | − | − | − |
| CCMP:1827 | Cryptoperidiniopsoid (S) | ? | − | − |
| CCMP:1828 | Cryptoperidiniopsoid (S) | ? | − | − |
| CCMP:1829 | Pfiesteria-like (L) | ? | − | − |
| CCMP:1830 | Pfiesteria-like (L) | ? | + | − |
| CCMP:1831 | Pfiesteria-like (L) | ? | + | − |
| CCMP:1832 | Pfiesteria-like (L) | ? | − | − |
| CCMP:1833 | Pfiesteria-like (L) | ? | − | − |
| CCMP:1834 | Pfiesteria-like (L) | ? | + | − |
| CCMP:1835 | Pfiesteria-like (L) | ? | − | − |
| CCMP:1836 | Pfiesteria-like (L) | ? | + | − |
Culture identification by scanning electron (S) or light (L) microscopy.
† FL DEP is culture MMRCC 981020BR01C5, identified as P. piscicida.
Scanning Electron Microscopy.
SEM characterization of dinoflagellate cultures (Fig. 1) was done as described (5, 7, 14). NCSU cultures Chic 97-1, 113-2, 114-1-5, 102-1, 97-1 and 125-4 as well as culture FL DEP MMRCC 981020BR01C5 were identified as P. piscicida. NCSU cultures BP, 7-28-T, and 15-T were identified as the provisional Pfiesteria species B.
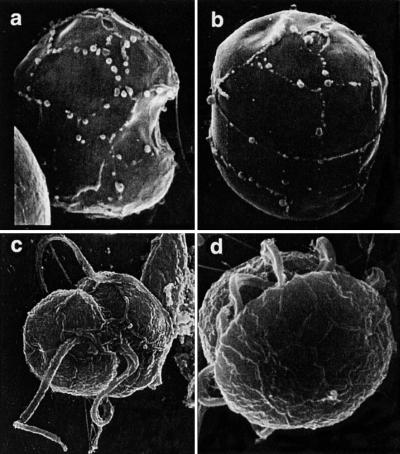
Figure 1.
Scanning electron micrographs of P. piscicida (a–c) and the related dinoflagellate, Pfiesteria species B (d). (a) P. piscicida culture MMRCC98-1020BR01C5 (Florida DEP) stripped of the outer wall layer. Sutures between plates clearly delineate X, 1′,2′, 4′, 1" 5" s.a., SD, and s.s. plates. The cell is 6.1 μm long. (b) Anterior dorsal view of stripped, swollen cell from same culture, revealing plates Po, Cp, X, 2–4′, 1a, and 2–4" in the epitheca. The cell is 7.3 μm long. The entire plate formula for this genus is Po, Cp, X, 4′, 1a, 5", 6c, 4s, 5′′′, 0p, and 2′′′. In addition to number of plates, position and shape are also important; note the small triangular 1a plate (arrow). (c) Ventral view, P. piscicida culture NCSU 113-2. Suture lines delineate 1′ and adjacent plates. (d) Pfiesteria species B culture NCSU 7-28-T; note the “diamond” (quadrangular as opposed to triangular) cingular plate.
Dinoflagellate DNA Extraction.
Estuarine water or culture samples were filtered (5-μm polyvinylidene filters; Millipore); the filters were then immersed in lysis buffer (DNeasy Plant Kit, Qiagen, Valencia CA) and were spun across a QIAShredder column (Qiagen), and DNA was eluted with buffer; thereafter, manufacturer's instructions were followed.
PCR Conditions.
Reactions (50 μl) contained 10–20 ng of template DNA, 50 mM KCl, 20 mM Tris⋅HCl (pH 8.4), 4.0 mM MgCl, 200 μM nucleotides, 1 unit of Taq DNA polymerase (PCR Supermix, GIBCO/Life Technologies, Gaithersburg, MD), and 0.8 μM of each specified primer. PCR conditions were as follows: for “dinoflagellate” PCR, primers dino/4618 (95°C, 3 min); then 40 cycles (95°C, 30 s/55°C, 30 s/72°C, 40 s); then 72°C, 5 min; for “species-specific” PCR, annealing temperature was 60°C. For longer reactions (primer pair Chic/4616) extension was increased to 60 s. PCR products were TA cloned into plasmid pCRR using a commercially available kit (TA CloningR Kit, Invitrogen). Plasmid minipreps were performed with a commercial kit (Qiagen). PCR primers: universal eukaryotic SSU 5′ primer 4616 (5′-AACCTGGTTGATCCTGCCAG-3′) and 3′ primer 4618 (5′-TGATCCTTCTG CAGGTTCACCTAC-3′) were adapted from ref. 14. “Dino” (5′-CGATTGAGTGATCCGGTGAATAA-3′). “Chic” (5′-AACTTTCCACTCCAACGTCCAG-3′). P. piscicida 108f (5′-AGTTAGATTGTCTTTGGTG GTCAA-3′). P. piscicida 311r (5′-GATAGGTCAGAAAGTGATATGGTA-3′). Species B-forward (5′-AGTTTTAGTGTA TTTGATGATCG-3′). Species B-reverse (5′-TCGAAAGCTGATAGGTCAGAATC-3′).
Combined Heteroduplex/Single Strand Conformational Polymorphism Assay.
Equivalent amounts of target and driver DNA were mixed (100 ng). Two hybridization strategies were utilized (Figs. 2 and 3); mixed PCR products were denatured at 95°C for 5 min, were cooled rapidly to 68°C, were maintained for 30 min, and then were slowly cooled (2°C/min) to room temperature (Fig. 4), or mixed PCR products were denatured at 95°C for 5 min and then were snap cooled on ice. Triple Dye loading buffer (FMC) was added to each tube. The samples were separated by electrophoresis across a 1× MDE gel (FMC) containing 4 M Urea, in a 20-cm Protein II xi electrophoresis unit (Bio-Rad) at 140 V over 24–36 hours [to optimize combined HMA/single-stranded conformational polymorphism (SSCP) data]. Gels were stained with SYBR Green II (FMC) for 30 min and were visualized on an Alpha Imager (Alpha Innotech, San Leandro, CA) utilizing a SYBR filter.
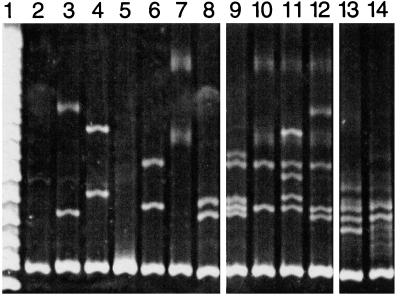
Figure 2.
Heteroduplex mobility assay (HMA) of dinoflagellate SSU rDNA sequences assayed against G. sanguineum “driver” DNA. Assayed “target” and driver DNAs were amplified with the primer pair (dino/4618); all lanes contain the same driver DNA, with added target sequences as indicated. Lanes: 1, DNA ladder; 2, G. sanguineum driver DNA alone; 3, Gymnodinium sp.; 4, P. minimum; 5, King's Creek, MD uncharacterized dinoflagellate DNA clone; 6, Pocomoke River, MD, uncharacterized dinoflagellate DNA clone; 7, uncharacterized dinoflagellate DNA clone derived from a lesioned menhaden (Brevoortia tyrannus); 8, P. piscicida; 9–12, intentionally mixed DNA(s) from lanes 3–8, demonstrating the ability of the assay to simultaneously detect multiple templates; 9, lanes 6 and 8; 10, lanes 6 and 7; 11, lanes 4, 6, and 7; 12, lanes 3, 4, 6, and 7 (the electrophoretic mobility of individual heteroduplex strands is not altered in the presence of mixed amplicons, but the overall HMA pattern become increasingly complex as potential strand annealing combinations multiply with increasing template DNA heterogeneity. With increasing complexity, some heteroduplex bands are lost); 13 and 14, dinoflagellate DNA derived from estuarine water samples (13, King's Creek, MD; 14, Pocomoke River, MD) demonstrating dinoflagellate diversity present in those samples at that time.
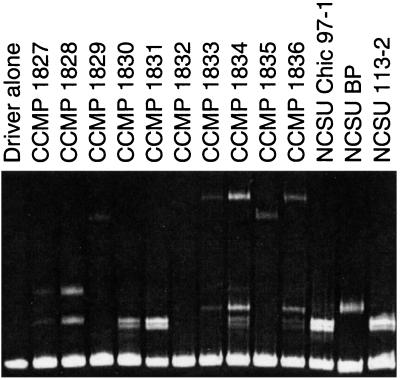
Figure 3.
HMA analysis of P. piscicida and pfiesteria-like dinoflagellate cultures. Individual lanes are as labeled; the driver sequence is as described in Fig. 2. HMA can be used to differentiate closely related dinoflagellate species (in this case, various isolates of pfiesteria-like dinoflagellates) and also to assess the clonality of dinoflagellate cultures (cultures CCMP 1834 and 1836 contained two distinct dinoflagellate sequences at this point in time).
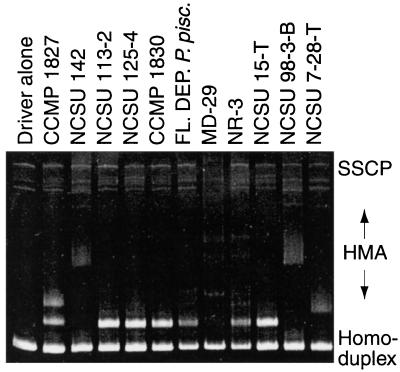
Figure 4.
Combined HMA/SSCP of dinoflagellate cultures and estuarine water samples. Lanes are as labeled; the driver DNA is as described in Fig. 2. Assayed specimens are described in Table 1. P. piscicida cultures established from different environmental isolates and in different laboratories demonstrate the same signature pattern. The same pattern is evident in assays of DNA extracted from filtered water samples collected from the Neuse River, NC in July, 1998 (NR-3) and the Chicamacomico River, MD, (MD-29) in September, 1997 during fish-kill events.
18S SSU DNA Sequence Alignment.
The alignment included 64 sequences, with an aligned length of 2,321 base pairs (338 characters were excluded from the analysis; the average sequence length was 1,770 bp, with a range of 1,650–1,940). Sequences were originally aligned with the GCG utility pileup (23) and were refined manually according to secondary structural information (24, 25). Phylogenetic analyses were performed with paup* 4.0d64 (26). The tree presented is a minimum evolution tree calculated with maximum likelihood distances using the GTR model, with invariant sites and gamma correction for site-to-site rate variation. All parameters were estimated, based on a preliminary tree calculated with Kimura 2 parameter distances, and the model selected was significantly better than simpler nested models when compared according to the likelihood ratio test (27). The same tree was found in 10 random addition sequence heuristic searches, suggesting that this is likely to be the best possible tree under the minimum evolution optimality criterion (28). (GenBank accession numbers are in Table 3 in the supplemental data)
Results
Dinoflagellate-Selective PCR.
An SSU sequence alignment including 40 dinoflagellate species and a broad selection of other eukaryotic taxa was created, and a primer sequence with selectivity for dinoflagellates was designed (dino). Used in conjunction with a “universal” 3′ eukaryotic 18S SSU primer (4618) and dinoflagellate template DNA, dino/4618 PCR produces a 142-bp fragment. Dino/4618 PCR did not amplify DNA from a variety of other estuarine plankton and metazoan species (Table 4 in the supplemental data) including, among the Alveolata Perkinsus marinus and multiple ciliate species. Conversely, this primer pair was able to amplify DNA from a broad sampling of dinoflagellate taxa including Prorocentrum minimum, Gonyaulax cochlea, Peridinium foliaceum, Gymnodinium sanguineum, and Coolia monotis (Table 2 in the supplemental data). Although the dino/4618 primer pair has a high degree of dinoflagellate selectivity, it is not absolute [Cryptosporidium sp. can also be amplified with this primer pair (data not shown)]. Through the course of field investigations conducted during 1998 and 1999 in Maryland and Delaware estuarine waters, DNA extracts from >500 filtered estuarine water samples have been assayed by PCR with the primer pair dino/4618, with rare negative results (Table 5 in the supplemental data).
HMA/SSCP Assays.
We performed mixing experiments with PCR-amplified DNA from characterized dinoflagellate cultures and/or cloned PCR products to assess the capability of HMA/SSCP to detect sequence heterogeneity in assayed samples. DNA amplified with primers dino/4618 from G. sanguineum was cloned (pGDINO) and reamplified as needed to produce the “driver” DNA utilized in these experiments. This PCR product migrates as a 142-bp homoduplex (Fig. 2, lane 2) when run alone in an MDE gel. Mixing of driver DNA with target DNA amplified from characterized cultures or cloned dino/4618 dinoflagellate PCR amplicons resulted in distinct heteroduplex patterns for five of six sequences (Fig. 2, lanes 3–8). On occasion, as seen in Fig. 2, lane 5, no heteroduplex bands are visualized. This may occur if driver and target DNA share 100% sequence identity or if marked sequence divergence prevents hybridization. In cases of sequence divergence, this difference is often detectable in the SSCP component of the gel (see Fig. 4). Two, three, or four distinct DNA sequences (in addition to driver DNA) could be detected simultaneously in intentional mixing experiments (Fig. 2, lanes 9–12). The HMA-detected sequence diversity among dino/4618 PCR amplicons from estuarine water sample DNA (Fig. 2, lanes 13 and 14) obtained from the Pocomoke Estuary and King's Creek (Manokin River), MD in the fall of 1997 during a time period of algal bloom and fish kill events. Definitive identification of sequences in complex hybridization mixtures in this context is, however, problematic, because, as the number of sequences increases, the number of possible hybridization events expands exponentially. Therefore, the assay was most useful for characterization of cultures and of panels of cloned PCR products.
To determine whether the HMA could differentiate dinoflagellates characterized at the light microscope level as “small heterotrophic estuarine dinoflagellates,” we next assayed a panel of CCMP dinoflagellate cultures (pfiesteria-like dinoflagellates) alongside organisms identified by SEM as P. piscicida (NCSU-Chic-97 and NCSU 113-2) and (provisionally) Pfiesteria species B (NCSU:BP) (Fig. 3). Among the 10 CCMP cultures, four distinct HMA patterns were observed. One culture, detected by primers 4618/dino, did not hybridize with the driver DNA (CCMP 1832). Two cultures believed to have been clonal for dinoflagellates were demonstrated to have at least two species of dinoflagellates present (CCMP 1834 and 1836). Subsequent clonal reisolates from these cultures segregated into two distinct HMA patterns (data not shown). Thus, the assay was of demonstrated utility in assessing the clonality of dinoflagellate cultures.
In Fig. 4, the reproducibility of these results is demonstrated. Three clonal cultures demonstrated to be P. piscicida by SEM (NCSU 113-2 and 125-4, and FL DEP P. piscicida) all demonstrate the same HMA pattern, as did CCMP 1830. Mixed (nonclonal for dinoflagellates) NCSU culture 15-T and environmental water sample NR-3 obtained from the Neuse Estuary, NC during a fish kill event in July, 1998 demonstrate the same distinctive pattern. Interestingly, included among the samples assayed in Fig. 4 are two associated with icthyotoxicity (cultures NCSU 113-2 and 125-4 by fish bioassay) as well as two others in which icthyotoxicity could not be demonstrated (FL DEP P. piscicida, CCMP 1830). 18S sequences derived from three of these cultures (FL DEP P. piscicida, CCMP 1830, and NCSU 113-2) are identical. Also reproduced in Fig. 4 is the distinct HMA pattern associated with Pfiesteria species B, visible in culture NCSU 7-28-T, and faintly in the mixed culture NCST 15-T (both P. piscicida and Pfiesteria species B patterns are visible). The SSCP patterns visible in Fig. 4 provide additional information; for instance, additional bands visible under SSCP for culture NCSU-142 suggested that this was a nonclonal culture, although only a single HMA band was visible. The dino/4618 PCR product from this culture was cloned, and 10 individual clones were run on HMA/SSCP, revealing two distinct SSCP patterns (data not shown).
During these investigations, a total of seven independently derived SEM confirmed P. piscicida cultures established at NCSU, all associated with icthyotoxicity in fish bioassays, were examined by HMA, and the same pattern was observed in each. Two clonal cultures characterized by SEM as (provisionally) Pfiesteria species B, each associated with icthyotoxicity in fish kill bioassays, were examined by HMA, and they shared a common HMA pattern. Thus, observed HMA signatures were consistent with SEM characterization across multiple cultures.
P. piscicida and Pfiesteria Species B 18S SSU rDNA Sequences.
As described above, it was possible to identify an HMA signature associated with multiple SEM confirmed P. piscicida cultures. The PCR amplicons generating this pattern were thus inferred to be derived from P. piscicida (in each of these cultures). Dino/4618 PCR products from these cultures were therefore cloned, sequenced, and found to be identical. A PCR primer targeted to this sequence, with maximal divergence from other dinoflagellate taxa, was designed (primer Chic). Near full length 18S gene PCR products from NCSU P. piscicida cultures 113-2 and Chic 97-1 (Table 1) were generated by using this primer in conjunction with a universal eukaryotic 5′ 18S gene primer (primer 4616). The distal 107-bp sequence (derived by sequencing the dino/4618 PCR product) was appended to generate the full length (1,807 bp) 18S SSU rDNA sequence for P. piscicida (GenBank accession no. AF077055). Further assurance that this sequence was derived from the authentic P. piscicida genome was provided by analysis of the panel of Pfiesteria cultures with HMA/SSCP, by species-specific PCR (described below), and by direct sequencing of full length PCR products from additional cultures in the panel.
Sequencing of the Pfiesteria species B 18S gene was also guided by HMA, although with a different cloning strategy. In this case, a culture believed to be clonal based on its' establishment through micromanipulation and the absence of dinoflagellate heterogeneity on HMA/SSCP assay was utilized (culture NCSU-BP). Full length 18S gene sequences were PCR amplified from the culture by utilizing the universal eukaryotic primers 4616/4618 and then were cloned and sequenced. A dinoflagellate sequence was obtained (GenBank accession no. AF218805). The linkage of this sequence with the provisional Pfiesteria species B was validated by confirmation that PCR with primers dino/4618 using the cloned DNA as template reproduced the previously observed HMA signature (data not shown), by sequencing of additional cultures, and by development of species-specific PCR for this sequence.
Species-Specific PCR Amplification of P. piscicida and Pfiesteria Species B.
By using the newly derived sequence data and the dinoflagellate 18S rDNA sequence matrix, P. piscicida-specific primers were designed (Ppisc108F/Ppisc311R). DNA extracted from each P. piscicida culture (identified by SEM and HMA) were amplified with this primer set, generating an appropriate size amplicons (200 bp). In addition, assays with the primer set Ppisc 108F/311R were positive with DNA extracted from environmental water samples and nonclonal cultures believed to contain P. piscicida, as presented in Table 1. This primer pair has high specificity for P. piscicida when used in a stringent assay, as suggested by multiple lines of evidence: (i) negative P. piscicida 108F/311R PCR assay results for more than 400 estuarine water samples (all testing positive with primers dino/4618) (Table 5 in the supplemental data); (ii) negative P. piscicida 108F/311R PCR assay results for 33 characterized non-Pfiesteria CCMP dinoflagellate cultures (Table 2 in the supplemental data) and all CCMP Pfiesteria-like dinoflagellate cultures that did not share the identical HMA pattern (n = 28; Table 2 in the supplemental data); (iii) negative P. piscicida 108F/311R PCR assay results for the most closely related dinoflagellate species available (Pfiesteria species B and Cryptoperidiniopsis species); and (iv) Sequence identity among all P. piscicida 108F/311R PCR product amplicons assayed to date (including those derived from environmental samples among which unidentified closely related organisms could be expected to occur).
In similar fashion, we designed PCR primers specific for the Pfiesteria species B 18S gene sequence (primers SpecB-forward/SpecB-reverse). This primer pair also appears to have high selectivity for the targeted gene sequence, based on specificity controls similar to those presented above (Table 2 in the supplemental data). However, in screening of cultures and environmental water samples, we have generated sequence data derived from at least one other uncharacterized organism (not present in our own or GenBank databases); we are further characterizing this sequence at this time. Thus, the Pfiesteria species B primer pair is highly selective but does not have absolute specificity for the targeted sequence (addition of a probe hybridization step enhances specificity, an area of further investigation in our labs) (data not shown).
Phylogenetic Position of Pfiesteria, and Other Aspects of the Phylogeny.
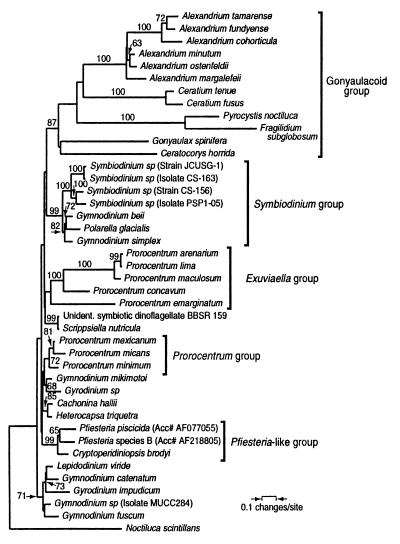
A preliminary analysis with a larger number of outgroup taxa placed the Pfiesteria sequences clearly among dinoflagellates (data not shown). To determine the relationship of P. piscicida and Pfiesteria species B to other dinoflagellates, we performed phylogenetic analyses on the full-length sequence alignment (Fig. 5). In most particulars, the phylogenetic analysis is consistent with previous analyses of SSU rDNA sequences of dinoflagellates (18). Although the ribosomal data alone do not determine a fully resolved tree, several noteworthy groups do find strong bootstrap support. The Pfiesteria-related clade composed of Pfiesteria piscicida, Pfiesteria species B, and Cryptoperidinopsis broydi has strong (99%) bootstrap support. Also strongly supported is a clade that includes species of Symbiodinium as well as Polarella glacialis and two other free-living species, Gymnodinium beii and Gymnodinium simplex. This clade is important both because it helps clarify the free-living relatives of Symbiodinium and because of the resemblance of Polarella cysts to the fossil Suessiales (29, 30). Also of interest is the division of the genus Prorocentrum into two distinct clades. These clades are consistent with morphological differences among the taxa and support the reinstatement of the genus Exuviaella (31, 32). Finally, gonyaulacoid and gymnodinioid groups also find moderate bootstrap support (87 and 71%, respectively), but the veracity of these clades awaits increased sampling of both taxa and gene sequence.
Figure 5.
Minimum evolution phylogenetic tree showing the placement of P. piscicida and Pfiesteria species B among dinoflagellates. P. piscicida (GenBank accession no. AF077055), Pfiesteria species B (GenBank accession no. AF218805), and C. brodyi (GenBank accession no. AF080097) cluster together in a well supported group (“Pfiesteria-like clade”) within dinoflagellates. Branching order within the dinoflagellates is not well resolved, although some groups have strong bootstrap support as described in text. Bootstrap values above 60% are indicated above the branches.
Discussion
Genetic sequence analysis is an invaluable tool in the study of dinoflagellate evolutionary systematics (18), and sequence-based methods have developed sufficiently to facilitate field and laboratory studies of microorganisms, even when present at low cell densities (33, 34). These approaches rely on the generation of sequence data, which for Pfiesteria and other heterotrophic dinoflagellates presented unique difficulties. One solution to this problem was the generation of “clonal” (single species dinoflagellate) cultures through single cell capillary tube transfer, as described above. Dinoflagellate-selective PCR amplification and HMA enhance those efforts by providing an assay to assess baseline and ongoing culture clonality. Alternatively, the HMA approach can be used to detect sequence signatures associated with culture or sample phenotypes of interest and subsequent derivation of full-length target gene sequences. The advantage of HMA-directed DNA sequence discovery in this context is its ability to identify specific sequences within complex assemblages or among nonculturable organisms. As presented in this manuscript, HMA can also be used as a tool to assign tentative identification to dinoflagellate cultures. Used in this manner, the assay performs similarly to restriction fragment length polymorphism analyses of PCR amplification products (35). Such an approach was recently deployed by Uribe et al. in comparing toxic Alexandrium cultures derived from Chilean isolates to reference cultures (36).
The value of HMA-guided sequence discovery was underscored by our own experience. During early sequencing efforts using universal SSU rDNA primers, we routinely cloned 18S gene sequences derived from algal prey (Cryptomonas sp., Rhodomonas sp.) and, in one case, a labyrinthuloides-like organism (data not shown). This experience, along with the demonstration that efforts to establish dinoflagellate clonality in cultures are not always successful (Figs. 3 and 4), convinced us that sequencing efforts needed to be guided by a selective strategy.
P. piscicida has been associated with fish morbidity and mortality in vitro (5, 11, 12), and the elaboration of a partially characterized toxin by the organism or an associate present in the fish-kill bioassay has been demonstrated (37). Fish placed in “active,” toxicity-associated cultures may rapidly develop aberrant behavior and epithelial hemorrhage (5, 11, 12, 38). However, the derivation of the incompletely characterized toxic substance in these cultures (i.e., toxin) is unknown. Use of sequence-based assays for identification of dinoflagellate species present in dinoflagellate/fish bioassays in conjunction with newly developed cell culture assays to detect toxicity (37) should enhance efforts to characterize these putative toxins. The observation that expression of toxic activity associated with P. piscicida cultures is variable raises intriguing questions for future investigations.
Heteroduplex analysis has been used to detect the presence of wild-type and mutant alleles in genetic counseling (20), to track the diversity and evolution of HIV and hepatitis C virus quasispecies in individual patients (39, 40), and to identify dinoflagellate strains (36) and has been proposed as a tool for broad molecular ecology applications (41). In this paper, we demonstrate an additional application of this versatile assay. HMA-directed sequence discovery is broadly applicable and may be adapted for the detection of genomic sequence data of novel or nonculturable organisms in complex assemblages.
Supplementary Material
Acknowledgments
We thank Nora Deamer, Elle Hannon, Alexandria Hamilton, and Zhang Cheng for cloning and maintaining Pfiesteria cultures (NCSU); Bill Richardson for providing clonal cultures of P. piscicida (FL DEP); Kathy Privett and Paula Scott for electron micrographs (FL DEP); Robert Anderson (Bigelow Laboratory) and contributors to the CCMP dinoflagellate culture panel; and Tsvetan Bachvaroff for assistance with analyses. Robert Magnien and David Goshorn of the Maryland Department of Natural Resources provided field samples. This work has been supported by the Maryland State Department of the Environment (D.W.O.), North Carolina Water Resources Research Institute (P.A.R.), Sea Grant Biotechnology Program (P.A.R.), the Environmental Protection Agency (D.W.O. and P.A.R.), Alfred P. Sloan Foundation Young Investigator Award 97-4-3 ME (C.F.D.), the Norwegian Research Council (K.S.J.), and Valborg Aschehougs Legat (T.T.). This is ECOHAB (Ecology and Oceanography of Harmful Algal Blooms) publication number 005.
Abbreviations
- HMA
heteroduplex mobility assay
- SSCP
single-stranded conformational polymorphism
- SEM
scanning electron microscopy
Footnotes
References
- 1.Harvell C D, Kim K, Burkholder J M, Colwell R R, Epstein P R, Grimes D J, Hofmann E E, Lipp E K, Osterhaus A D M E, Overstreet R M, et al. Science. 1999;285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- 2.Hallegraeff G M. Phycologia. 1993;32:79–99. [Google Scholar]
- 3.Smayda T J. In: Toxic Marine Phytoplankton. Graneli E, Sundstrom B, Edler L, Anderson D M, editors. New York: Elsevier Science; 1996. pp. 29–40. [Google Scholar]
- 4.Burkholder J M, Glasgow H B., Jr Arch Protistenk. 1995;145:177–188. [Google Scholar]
- 5.Burkholder J M, Noga E J, Hobbs C W, Glasgow H B, Jr, Smith S A. Nature (London) 1992;358:407–410. doi: 10.1038/358407a0. , and erratum (1992) 360, 768. [DOI] [PubMed] [Google Scholar]
- 6.Steidinger K A, Burkholder J M, Glasgow H B, Jr, Truby E, Garrett J, Noga E J, Smith S A. J Phycol. 1996;32:157–164. [Google Scholar]
- 7.Burkholder J M, Glasgow H B, Jr, Hobbs C W. Mar Ecol Prog Ser. 1995;124:43–61. [Google Scholar]
- 8.Glasgow H B, Jr, Burkholder J M, Schmechel D E, Tester P A. J Toxicol Environ Health. 1995;46:501–522. doi: 10.1080/15287399509532051. [DOI] [PubMed] [Google Scholar]
- 9.Grattan L M, Oldach D W, Perl T M, Lowitt M H, Matuszak D L, Dickson C, Parrott C, Shoemaker R C, Kauffman C L, Wasserman M P, et al. Lancet. 1998;352:532–539. doi: 10.1016/S0140-6736(98)02132-1. [DOI] [PubMed] [Google Scholar]
- 10.Oldach D W, Grattan L M, Morris J G., Jr . In: Emerging Infections 3. Scheld W M, Craig W A, Hughes J M, editors. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 135–151. [Google Scholar]
- 11.Burkholder J M, Glasgow H B., Jr J Eukaryot Microbiol. 1997;44:200–205. doi: 10.1111/j.1550-7408.1997.tb05700.x. [DOI] [PubMed] [Google Scholar]
- 12.Burkholder J M, Glasgow H B., Jr Limnol Oceanogr. 1997;42:1052–1075. [Google Scholar]
- 13.Steidinger K A, Landsberg J H, Truby E W, Blakesly B A. Nova Hedwigia. 1996;112:415–422. [Google Scholar]
- 14.Medlin L, Elwood H J, Stickel S, Sogin M L. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 15.Sogin M L, Gunderson J M. Ann NY Acad Sci. 1987;503:125–139. doi: 10.1111/j.1749-6632.1987.tb40603.x. [DOI] [PubMed] [Google Scholar]
- 16.Elwood H J, Olsen G J, Sogin M L. Mol Biol Evol. 1985;2:399–410. doi: 10.1093/oxfordjournals.molbev.a040362. [DOI] [PubMed] [Google Scholar]
- 17.Sogin M L. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 307–314. [Google Scholar]
- 18.Saunders G W, Hill D R A, Sexton J P, Andersen R A. Plant Syst Evol. 1997;11,Suppl.:237–259. [Google Scholar]
- 19.Maidak B L, Olsen G J, Larsen, Overbeck R, McCaughey M J, Woese C R. Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glavac D, Dean M. In: Technologies for Detection of DNA Damage and Mutations. Pfeifer G, editor. NY: Plenum; 1997. pp. 241–251. [Google Scholar]
- 21.Nielsen M V. Mar Ecol Prog Ser. 1993;95:273–277. [Google Scholar]
- 22.Noga E J, Bower C E. J Parasitol. 1987;73:924–928. [PubMed] [Google Scholar]
- 23.Genetics Computer Group. wisconsin package 9.1. Madison, WI: Genetics Computer Group; 1997. [Google Scholar]
- 24.Gutell R R. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van de Peer Y, Caers A, De Rijk P, De Wachter R. Nucleic Acids Res. 1998;26:179–182. [PMC free article] [PubMed] [Google Scholar]
- 26.Swofford D L. paup*: Phylogenetic Analysis Using Parsimony. Sunderland, MA: Sinauer; 1999. [Google Scholar]
- 27.Huelsenbeck J P, Rannala B. Science. 1997;276:227–232. doi: 10.1126/science.276.5310.227. [DOI] [PubMed] [Google Scholar]
- 28.Swofford D L, Olsen G J, Waddell P J, Hillis D M. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 407–514. [Google Scholar]
- 29.Montresor M, Procaccini G, Stoecker D K. J Phycol. 1999;35:186–197. [Google Scholar]
- 30.Carlos A A, Baillie B K, Kawachi M, Maruyama T. J Phycol. 1999;35:1054–1062. [Google Scholar]
- 31.McLachlan J L, Boalch G T, Jahn R. Phycologia. 1997;36:38–46. [Google Scholar]
- 32.Grzebyk D, Sako Y, Berland B. J Phycol. 1998;34:1055–1068. [Google Scholar]
- 33.Barnes S M, Delwiche C F, Palmer J D, Pace N R. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholin C A, Anderson D M. J Phycol. 1994;30:744–754. [Google Scholar]
- 35.Scholin C A, Hallegraeff G M, Anderson D M. Phycologia. 1995;34:472–485. [Google Scholar]
- 36.Uribe P, Suarez-Isla B A, Espejo R T. J Phycol. 1999;35:884–888. [Google Scholar]
- 37.Fairey E R, Edmunds J S G, Deamer-Melia N J, Glasgow H B, Jr, Johnson F M, Moeller P R, Burkholder J M, Ramsdell J S. Environ Health Perspect. 1999;107:711–714. doi: 10.1289/ehp.99107711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noga E J, Khoo L, Stevens J B, Fan Z, Burkholder J M. Mar Pollution Bull. 1996;32:219–224. [Google Scholar]
- 39.Nelson J A, Fiscus S A, Swanstrom R. J Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y M, Ray S C, Laeyendecker O, Ticehurst J R, Thomas D L. J Clin Microbiol. 1998;36:2982–2989. doi: 10.1128/jcm.36.10.2982-2989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lessa E P, Applebaum G. Mol Ecol. 1993;2:119–129. doi: 10.1111/j.1365-294x.1993.tb00006.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.