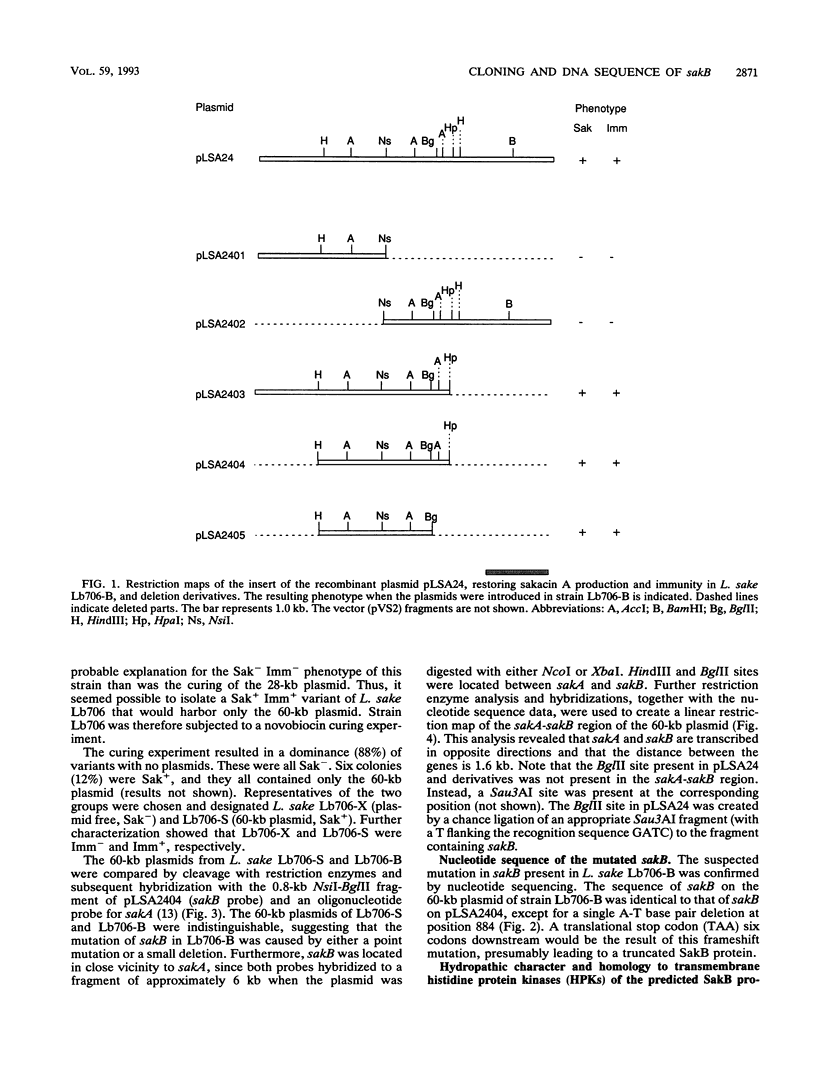
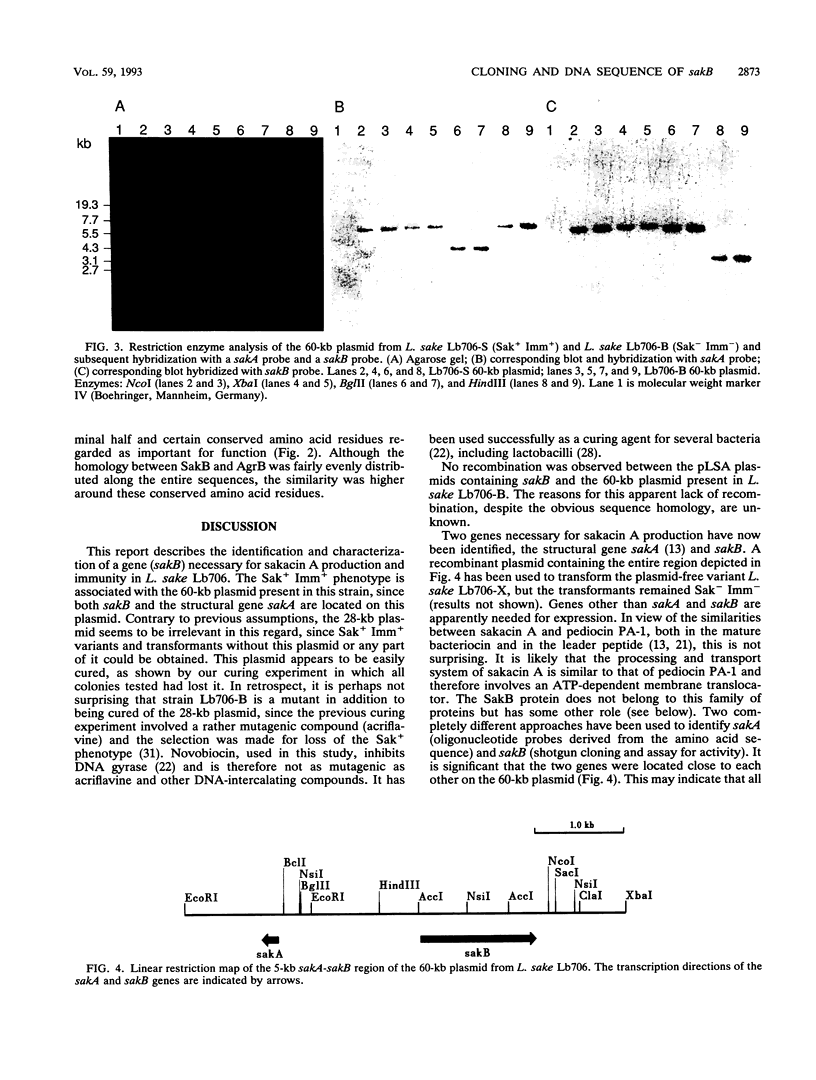
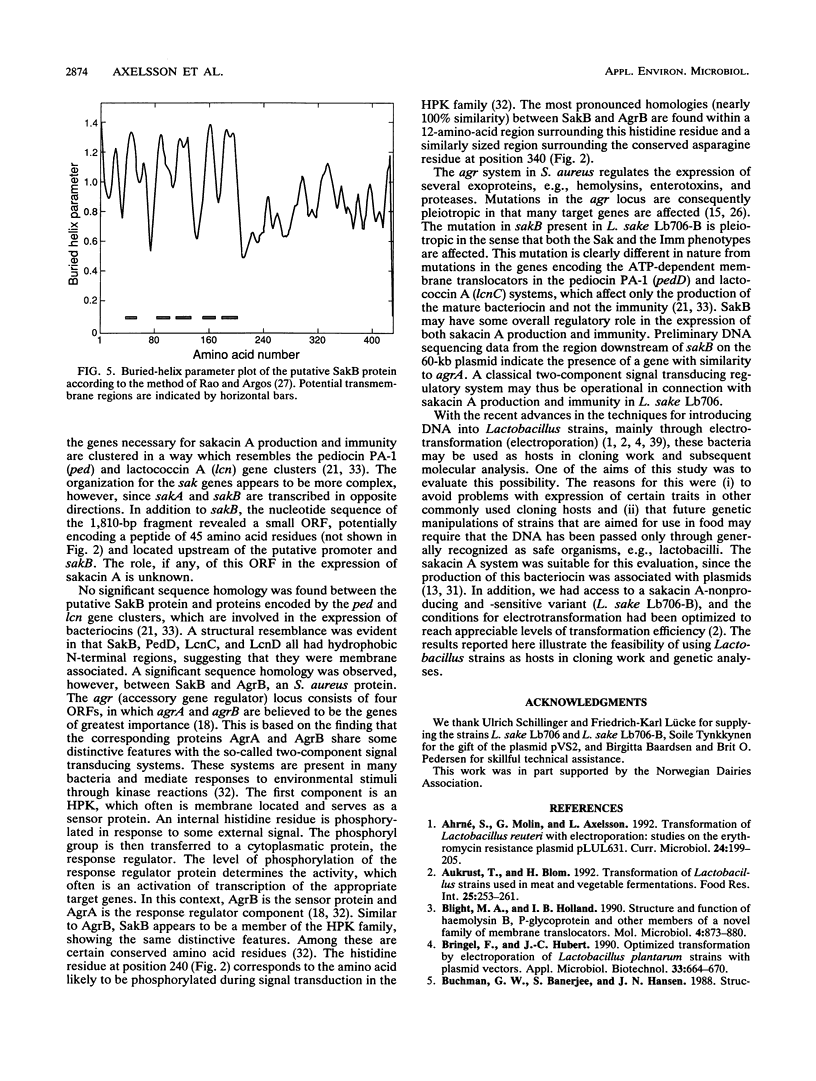
Abstract
Sakacin A is an antilisterial bacteriocin produced by Lactobacillus sake Lb706. In order to identify genes involved in sakacin A production and immunity, the plasmid fraction of L. sake Lb706 was shotgun cloned directly into a sakacin A-nonproducing and -sensitive variant, L. sake Lb706-B, by using the broad-host-range vector pVS2. Two clones that produced sakacin A and were immune to the bacteriocin were obtained. A DNA fragment of approximately 1.8 kb, derived from a 60-kb plasmid of strain Lb706 and present in the inserts of both clones, was necessary for restoration of sakacin A production and immunity in strain Lb706-B. The sequence of the 1.8-kb fragment from one of the clones was determined. It contained one large open reading frame, designated sakB, potentially encoding a protein of 430 amino acid residues. Hybridization and nucleotide sequence analyses revealed that the cloned sakB complemented a mutated copy of sakB present in strain Lb706-B. The sakB gene mapped 1.6 kb from the previously cloned structural gene for sakacin A (sakA) on the 60-kb plasmid. The putative SakB protein shared 22% amino acid sequence identity (51% similarity if conservative changes are considered) to AgrB, the deduced amino acid sequence of the Staphylococcus aureus gene agrB. The polycistronic agr (accessory gene regulator) locus is involved in the regulation of exoprotein synthesis in S. aureus. Similar to the AgrB protein, SakB had some features in common with a family of transmembrane histidine protein kinases, involved in various adaptive response systems of bacteria.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blight M. A., Holland I. B. Structure and function of haemolysin B,P-glycoprotein and other members of a novel family of membrane translocators. Mol Microbiol. 1990 Jun;4(6):873–880. doi: 10.1111/j.1365-2958.1990.tb00660.x. [DOI] [PubMed] [Google Scholar]
- Buchman G. W., Banerjee S., Hansen J. N. Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J Biol Chem. 1988 Nov 5;263(31):16260–16266. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke G., Gutowski-Eckel Z., Hammelmann M., Entian K. D. Biosynthesis of the lantibiotic nisin: genomic organization and membrane localization of the NisB protein. Appl Environ Microbiol. 1992 Nov;58(11):3730–3743. doi: 10.1128/aem.58.11.3730-3743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves M. C., Rabinowitz J. C. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. Evidence for "extended" promoter elements in gram-positive organisms. J Biol Chem. 1986 Aug 25;261(24):11409–11415. [PubMed] [Google Scholar]
- Gross E., Morell J. L. The structure of nisin. J Am Chem Soc. 1971 Sep 8;93(18):4634–4635. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- Hastings J. W., Sailer M., Johnson K., Roy K. L., Vederas J. C., Stiles M. E. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J Bacteriol. 1991 Dec;173(23):7491–7500. doi: 10.1128/jb.173.23.7491-7500.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holck A., Axelsson L., Birkeland S. E., Aukrust T., Blom H. Purification and amino acid sequence of sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Gen Microbiol. 1992 Dec;138(12):2715–2720. doi: 10.1099/00221287-138-12-2715. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzon L., Löfdahl S., Arvidson S. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol Gen Genet. 1989 Nov;219(3):480–485. doi: 10.1007/BF00259623. [DOI] [PubMed] [Google Scholar]
- Kaletta C., Entian K. D. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J Bacteriol. 1989 Mar;171(3):1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R. Bacteriocins of lactic acid bacteria. Biochimie. 1988 Mar;70(3):337–349. doi: 10.1016/0300-9084(88)90206-4. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R., Tardiff J., Krauter K. S., Leinwand L. A. Using mini-prep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques. 1988 Jun;6(6):544-6, 549. [PubMed] [Google Scholar]
- Marugg J. D., Gonzalez C. F., Kunka B. S., Ledeboer A. M., Pucci M. J., Toonen M. Y., Walker S. A., Zoetmulder L. C., Vandenbergh P. A. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, and bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992 Aug;58(8):2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh G. L., Swartz M. N. Elimination of plasmids from several bacterial species by novobiocin. Antimicrob Agents Chemother. 1977 Sep;12(3):423–426. doi: 10.1128/aac.12.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Applications for biotechnology: present and future improvements in lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):3–14. doi: 10.1111/j.1574-6968.1990.tb04876.x. [DOI] [PubMed] [Google Scholar]
- Mohana Rao J. K., Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986 Jan 30;869(2):197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- Muriana P. M., Klaenhammer T. R. Cloning, phenotypic expression, and DNA sequence of the gene for lactacin F, an antimicrobial peptide produced by Lactobacillus spp. J Bacteriol. 1991 Mar;173(5):1779–1788. doi: 10.1128/jb.173.5.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto Lozano J. C., Meyer J. N., Sletten K., Peláz C., Nes I. F. Purification and amino acid sequence of a bacteriocin produced by Pediococcus acidilactici. J Gen Microbiol. 1992 Sep;138(9):1985–1990. doi: 10.1099/00221287-138-9-1985. [DOI] [PubMed] [Google Scholar]
- Peng H. L., Novick R. P., Kreiswirth B., Kornblum J., Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988 Sep;170(9):4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Barba J. L., Piard J. C., Jiménez-Díaz R. Plasmid profiles and curing of plasmids in Lactobacillus plantarum strains isolated from green olive fermentations. J Appl Bacteriol. 1991 Nov;71(5):417–421. doi: 10.1111/j.1365-2672.1991.tb03810.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillinger U., Lücke F. K. Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol. 1989 Aug;55(8):1901–1906. doi: 10.1128/aem.55.8.1901-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard G. W., Petzel J. P., van Belkum M. J., Kok J., McKay L. L. Molecular analyses of the lactococcin A gene cluster from Lactococcus lactis subsp. lactis biovar diacetylactis WM4. Appl Environ Microbiol. 1992 Jun;58(6):1952–1961. doi: 10.1128/aem.58.6.1952-1961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagg J. R., Dajani A. S., Wannamaker L. W. Bacteriocins of gram-positive bacteria. Bacteriol Rev. 1976 Sep;40(3):722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch F., Kornblum J., Novick R. P. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J Bacteriol. 1991 Oct;173(20):6313–6320. doi: 10.1128/jb.173.20.6313-6320.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wright A., Tynkkynen S., Suominen M. Cloning of a Streptococcus lactis subsp. lactis Chromosomal Fragment Associated with the Ability To Grow in Milk. Appl Environ Microbiol. 1987 Jul;53(7):1584–1588. doi: 10.1128/aem.53.7.1584-1588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]




