Abstract
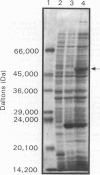
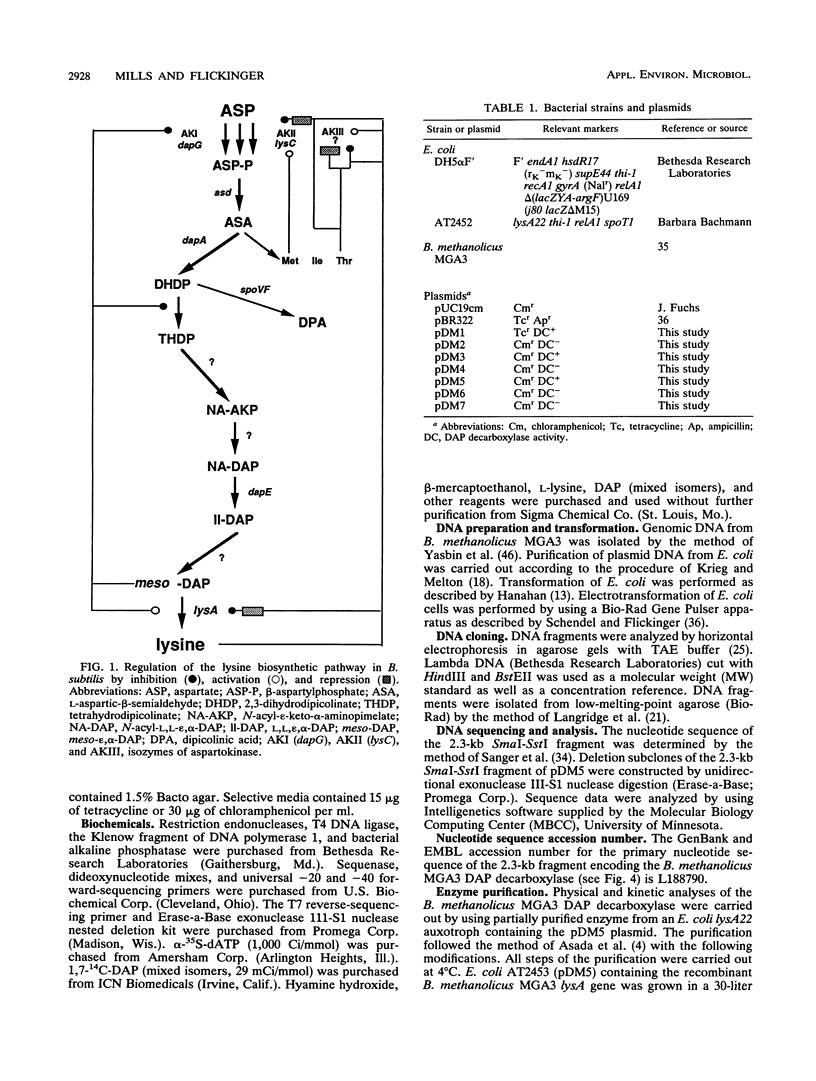
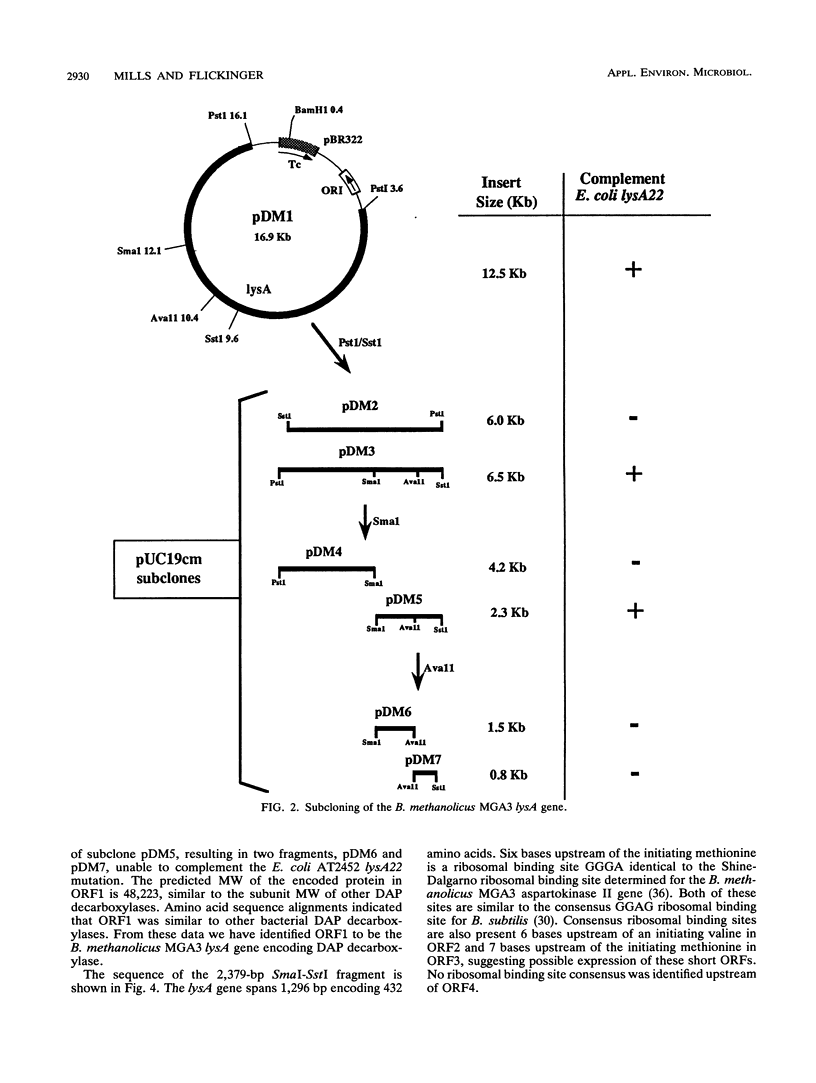
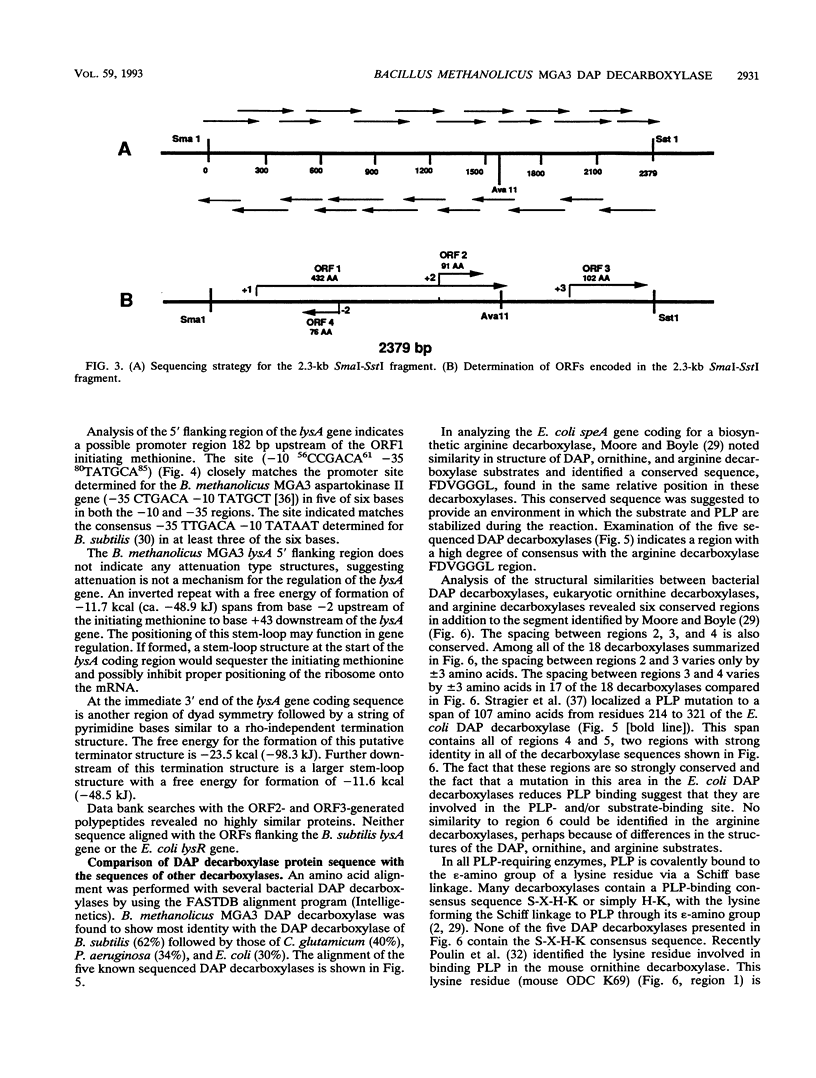
The lysA gene of Bacillus methanolicus MGA3 was cloned by complementation of an auxotrophic Escherichia coli lysA22 mutant with a genomic library of B. methanolicus MGA3 chromosomal DNA. Subcloning localized the B. methanolicus MGA3 lysA gene into a 2.3-kb SmaI-SstI fragment. Sequence analysis of the 2.3-kb fragment indicated an open reading frame encoding a protein of 48,223 Da, which was similar to the meso-diaminopimelate (DAP) decarboxylase amino acid sequences of Bacillus subtilis (62%) and Corynebacterium glutamicum (40%). Amino acid sequence analysis indicated several regions of conservation among bacterial DAP decarboxylases, eukaryotic ornithine decarboxylases, and arginine decarboxylases, suggesting a common structural arrangement for positioning of substrate and the cofactor pyridoxal 5'-phosphate. The B. methanolicus MGA3 DAP decarboxylase was shown to be a dimer (M(r) 86,000) with a subunit molecular mass of approximately 50,000 Da. This decarboxylase is inhibited by lysine (Ki = 0.93 mM) with a Km of 0.8 mM for DAP. The inhibition pattern suggests that the activity of this enzyme in lysine-overproducing strains of B. methanolicus MGA3 may limit lysine synthesis.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applebaum D. M., Dunlap J. C., Morris D. R. Comparison of the biosynthetic and biodegradative ornithine decarboxylases of Escherichia coli. Biochemistry. 1977 Apr 19;16(8):1580–1584. doi: 10.1021/bi00627a008. [DOI] [PubMed] [Google Scholar]
- Arfman N., Dijkhuizen L., Kirchhof G., Ludwig W., Schleifer K. H., Bulygina E. S., Chumakov K. M., Govorukhina N. I., Trotsenko Y. A., White D. Bacillus methanolicus sp. nov., a new species of thermotolerant, methanol-utilizing, endospore-forming bacteria. Int J Syst Bacteriol. 1992 Jul;42(3):439–445. doi: 10.1099/00207713-42-3-439. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I., Taylor A. L. Mutants of Escherichia coli with a growth requirement for either lysine or pyridoxine. J Bacteriol. 1971 Mar;105(3):988–998. doi: 10.1128/jb.105.3.988-998.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland W. W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- Cremer J., Treptow C., Eggeling L., Sahm H. Regulation of enzymes of lysine biosynthesis in Corynebacterium glutamicum. J Gen Microbiol. 1988 Dec;134(12):3221–3229. doi: 10.1099/00221287-134-12-3221. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Stahly D. P. Repression of diaminopimelate decarboxylase by L-lysine in different Bacillus species. J Bacteriol. 1971 Mar;105(3):1211–1212. doi: 10.1128/jb.105.3.1211-1212.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves L. M., Switzer R. L. Aspartokinase III, a new isozyme in Bacillus subtilis 168. J Bacteriol. 1990 Jan;172(1):218–223. doi: 10.1128/jb.172.1.218-223.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelland J. G., Palcic M. M., Pickard M. A., Vederas J. C. Stereochemistry of lysine formation by meso-diaminopimelate decarboxylase from wheat germ: use of 1H-13C NMR shift correlation to detect stereospecific deuterium labeling. Biochemistry. 1985 Jun 18;24(13):3263–3267. doi: 10.1021/bi00334a028. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langridge J., Langridge P., Bergquist P. L. Extraction of nucleic acids from agarose gels. Anal Biochem. 1980 Apr;103(2):264–271. doi: 10.1016/0003-2697(80)90266-3. [DOI] [PubMed] [Google Scholar]
- Lidstrom M. E., Tsygankov Y. D. Molecular genetics of methylotrophic bacteria. Biotechnology. 1991;18:273–304. doi: 10.1016/b978-0-7506-9188-8.50019-x. [DOI] [PubMed] [Google Scholar]
- Lu L., Stanley B. A., Pegg A. E. Identification of residues in ornithine decarboxylase essential for enzymic activity and for rapid protein turnover. Biochem J. 1991 Aug 1;277(Pt 3):671–675. doi: 10.1042/bj2770671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel T., Archer J. A., Mengin-Lecreulx D., Sinskey A. J. Nucleotide sequence and organization of the upstream region of the Corynebacterium glutamicum lysA gene. Mol Microbiol. 1990 Nov;4(11):1819–1830. doi: 10.1111/j.1365-2958.1990.tb02030.x. [DOI] [PubMed] [Google Scholar]
- Martin C., Cami B., Yeh P., Stragier P., Parsot C., Patte J. C. Pseudomonas aeruginosa diaminopimelate decarboxylase: evolutionary relationship with other amino acid decarboxylases. Mol Biol Evol. 1988 Sep;5(5):549–559. doi: 10.1093/oxfordjournals.molbev.a040515. [DOI] [PubMed] [Google Scholar]
- Moore R. C., Boyle S. M. Nucleotide sequence and analysis of the speA gene encoding biosynthetic arginine decarboxylase in Escherichia coli. J Bacteriol. 1990 Aug;172(8):4631–4640. doi: 10.1128/jb.172.8.4631-4640.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Márquez G., Sousa J. M., Sánchez F. Cloning and expression in Escherichia coli of genes involved in the lysine pathway of Brevibacterium lactofermentum. J Bacteriol. 1985 Oct;164(1):379–383. doi: 10.1128/jb.164.1.379-383.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Alterthum F., Ingram L. O. Effects of environmental conditions on xylose fermentation by recombinant Escherichia coli. Appl Environ Microbiol. 1990 Feb;56(2):463–465. doi: 10.1128/aem.56.2.463-465.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin R., Lu L., Ackermann B., Bey P., Pegg A. E. Mechanism of the irreversible inactivation of mouse ornithine decarboxylase by alpha-difluoromethylornithine. Characterization of sequences at the inhibitor and coenzyme binding sites. J Biol Chem. 1992 Jan 5;267(1):150–158. [PubMed] [Google Scholar]
- Rosner A. Control of lysine biosynthesis in Bacillus subtilis: inhibition of diaminopimelate decarboxylase by lysine. J Bacteriol. 1975 Jan;121(1):20–28. doi: 10.1128/jb.121.1.20-28.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendel F. J., Flickinger M. C. Cloning and nucleotide sequence of the gene coding for aspartokinase II from a thermophilic methylotrophic Bacillus sp. Appl Environ Microbiol. 1992 Sep;58(9):2806–2814. doi: 10.1128/aem.58.9.2806-2814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Danos O., Patte J. C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. II. Nucleotide sequence of the lysA gene and its regulatory region. J Mol Biol. 1983 Aug 5;168(2):321–331. doi: 10.1016/s0022-2836(83)80021-7. [DOI] [PubMed] [Google Scholar]
- Stragier P., Patte J. C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. III. Nucleotide sequence and regulation of the lysR gene. J Mol Biol. 1983 Aug 5;168(2):333–350. doi: 10.1016/s0022-2836(83)80022-9. [DOI] [PubMed] [Google Scholar]
- Stragier P., Richaud F., Borne F., Patte J. C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. I. Identification of a lysR gene encoding an activator of the lysA gene. J Mol Biol. 1983 Aug 5;168(2):307–320. doi: 10.1016/s0022-2836(83)80020-5. [DOI] [PubMed] [Google Scholar]
- WHITE P. J., KELLY B. PURIFICATION AND PROPERTIES OF DIAMINOPIMELATE DECARBOXYLASE FROM ESCHERICHIA COLI. Biochem J. 1965 Jul;96:75–84. doi: 10.1042/bj0960075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N., Dunyak D. S., Rosey E. L., Slonczewski J. L., Olson E. R. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992 Jan;174(2):530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto J., Shimizu M., Yamane K. Molecular cloning and analysis of nucleotide sequence of the Bacillus subtilis lysA gene region using B. subtilis phage vectors and a multi-copy plasmid, pUB110. Agric Biol Chem. 1991 Jun;55(6):1615–1626. [PubMed] [Google Scholar]
- Yamamoto J., Shimizu M., Yamane K. Nucleotide sequence of the diaminopimelate-decarboxylase gene from Bacillus subtilis. Nucleic Acids Res. 1989 Dec 11;17(23):10105–10105. doi: 10.1093/nar/17.23.10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasbin R. E., Wilson G. A., Young F. E. Transformation and transfection in lysogenic strains of Bacillus subtilis: evidence for selective induction of prophage in competent cells. J Bacteriol. 1975 Jan;121(1):296–304. doi: 10.1128/jb.121.1.296-304.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh P., Sicard A. M., Sinskey A. J. Nucleotide sequence of the lysA gene of Corynebacterium glutamicum and possible mechanisms for modulation of its expression. Mol Gen Genet. 1988 Apr;212(1):112–119. doi: 10.1007/BF00322452. [DOI] [PubMed] [Google Scholar]
- de Vries G. E., Arfman N., Terpstra P., Dijkhuizen L. Cloning, expression, and sequence analysis of the Bacillus methanolicus C1 methanol dehydrogenase gene. J Bacteriol. 1992 Aug;174(16):5346–5353. doi: 10.1128/jb.174.16.5346-5353.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]