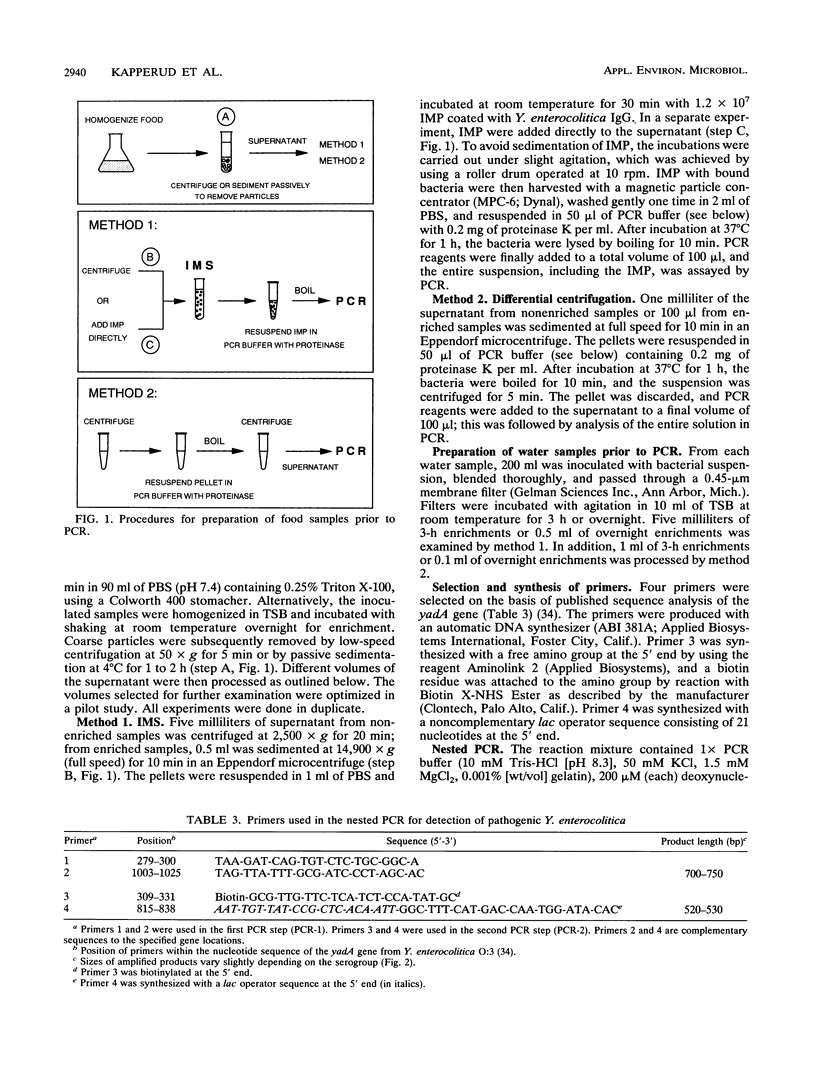
Abstract
A two-step polymerase chain reaction (PCR) procedure with two nested pairs of primers specific for the yadA gene of Yersinia enterocolitica was developed. The PCR assay identified all common pathogenic serogroups (O:3, O:5,27, O:8, O:9, O:13, and O:21) from three continents and differentiated pathogenic Y. enterocolitica from Y. pseudotuberculosis and from a variety of nonpathogenic yersiniae representing 25 serogroups and four species. The performance of the method was evaluated with seeded food and water samples. We compared two procedures for sample preparation prior to PCR: one was based on immunomagnetic separation of the target bacteria from the sample, using magnetic particles coated with immunoglobulin antibodies to Y. enterocolitica serogroup O:3, and the other method consisted of a series of centrifugation steps combined with proteinase treatment. Regardless of the method used, the PCR assay was capable of detecting 10 to 30 CFU/g of meat in 10(6)-fold excess of indigenous bacteria. When the samples were enriched overnight in a nonselective medium, the sensitivity was increased to approximately 2 CFU/g, except for samples with an extremely high background flora (> 10(7) CFU/g). We compared gel electrophoretic detection of PCR products with a colorimetric detection method designated DIANA (detection of immobilized amplified nucleic acids), which enabled easy visualization of amplified fragments in a microtiter plate format with an optical density reader. DIANA and gel electrophoresis showed complete concordance in their discrimination between positive and negative samples. The combination of immunomagnetic separation, nested PCR, and DIANA makes possible the development of a fully automated analytic process which requires a minimum of laboratory manipulations.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Christensen B., Torsvik T., Lien T. Immunomagnetically captured thermophilic sulfate-reducing bacteria from north sea oil field waters. Appl Environ Microbiol. 1992 Apr;58(4):1244–1248. doi: 10.1128/aem.58.4.1244-1248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover T. L., Aber R. C. Yersinia enterocolitica. N Engl J Med. 1989 Jul 6;321(1):16–24. doi: 10.1056/NEJM198907063210104. [DOI] [PubMed] [Google Scholar]
- Feng P., Keasler S. P., Hill W. E. Direct identification of Yersinia enterocolitica in blood by polymerase chain reaction amplification. Transfusion. 1992 Nov-Dec;32(9):850–854. doi: 10.1046/j.1537-2995.1992.32993110759.x. [DOI] [PubMed] [Google Scholar]
- Fenwick S. G., Murray A. Detection of pathogenic Yersinia enterocolitica by polymerase chain reaction. Lancet. 1991 Feb 23;337(8739):496–497. doi: 10.1016/0140-6736(91)93436-d. [DOI] [PubMed] [Google Scholar]
- Hornes E., Wasteson Y., Olsvik O. Detection of Escherichia coli heat-stable enterotoxin genes in pig stool specimens by an immobilized, colorimetric, nested polymerase chain reaction. J Clin Microbiol. 1991 Nov;29(11):2375–2379. doi: 10.1128/jcm.29.11.2375-2379.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman T., Ståhl S., Hornes E., Uhlén M. Direct solid phase sequencing of genomic and plasmid DNA using magnetic beads as solid support. Nucleic Acids Res. 1989 Jul 11;17(13):4937–4946. doi: 10.1093/nar/17.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A., Liesack W., Stackebrandt E. Polymerase chain reaction-gene probe detection system specific for pathogenic strains of Yersinia enterocolitica. J Clin Microbiol. 1992 Aug;30(8):1942–1947. doi: 10.1128/jcm.30.8.1942-1947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Feinstone S. M., Miller R. H. Rapid and sensitive method for the detection of serum hepatitis B virus DNA using the polymerase chain reaction technique. J Clin Microbiol. 1989 Sep;27(9):1930–1933. doi: 10.1128/jcm.27.9.1930-1933.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapperud G., Namork E., Skurnik M., Nesbakken T. Plasmid-mediated surface fibrillae of Yersinia pseudotuberculosis and Yersinia enterocolitica: relationship to the outer membrane protein YOP1 and possible importance for pathogenesis. Infect Immun. 1987 Sep;55(9):2247–2254. doi: 10.1128/iai.55.9.2247-2254.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapperud G., Nesbakken T., Aleksic S., Mollaret H. H. Comparison of restriction endonuclease analysis and phenotypic typing methods for differentiation of Yersinia enterocolitica isolates. J Clin Microbiol. 1990 Jun;28(6):1125–1131. doi: 10.1128/jcm.28.6.1125-1131.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapperud G. Yersinia enterocolitica in food hygiene. Int J Food Microbiol. 1991 Jan;12(1):53–65. doi: 10.1016/0168-1605(91)90047-s. [DOI] [PubMed] [Google Scholar]
- Kemp D. J., Smith D. B., Foote S. J., Samaras N., Peterson M. G. Colorimetric detection of specific DNA segments amplified by polymerase chain reactions. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2423–2427. doi: 10.1073/pnas.86.7.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaga J., Iversen J. O., Misra V. Detection of pathogenic Yersinia enterocolitica by polymerase chain reaction and digoxigenin-labeled polynucleotide probes. J Clin Microbiol. 1992 Oct;30(10):2668–2673. doi: 10.1128/jcm.30.10.2668-2673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk J. M., Lindberg A. A. Rapid and sensitive detection of Salmonella (O:6,7) by immunomagnetic monoclonal antibody-based assays. J Immunol Methods. 1991 Mar 1;137(1):1–8. doi: 10.1016/0022-1759(91)90387-u. [DOI] [PubMed] [Google Scholar]
- Lund A., Hellemann A. L., Vartdal F. Rapid isolation of K88+ Escherichia coli by using immunomagnetic particles. J Clin Microbiol. 1988 Dec;26(12):2572–2575. doi: 10.1128/jcm.26.12.2572-2575.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollaret H. H., Bercovier H., Alonso J. M. Summary of the data received at the WHO Reference Center for Yersinia enterocolitica. Contrib Microbiol Immunol. 1979;5:174–184. [PubMed] [Google Scholar]
- Morgan J. A., Winstanley C., Pickup R. W., Saunders J. R. Rapid Immunocapture of Pseudomonas putida Cells from Lake Water by Using Bacterial Flagella. Appl Environ Microbiol. 1991 Feb;57(2):503–509. doi: 10.1128/aem.57.2.503-509.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H., Inoue M., Mori T., Itoh K., Arakawa E., Watanabe H. Detection and identification of Yersinia pseudotuberculosis and pathogenic Yersinia enterocolitica by an improved polymerase chain reaction method. J Clin Microbiol. 1992 Sep;30(9):2484–2486. doi: 10.1128/jcm.30.9.2484-2486.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbakken T., Kapperud G., Dommarsnes K., Skurnik M., Hornes E. Comparative study of a DNA hybridization method and two isolation procedures for detection of Yersinia enterocolitica O:3 in naturally contaminated pork products. Appl Environ Microbiol. 1991 Feb;57(2):389–394. doi: 10.1128/aem.57.2.389-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbakken T., Kapperud G., Sørum H., Dommarsnes K. Structural variability of 40-50 Mdal virulence plasmids from Yersinia enterocolitica. Geographical and ecological distribution of plasmid variants. Acta Pathol Microbiol Immunol Scand B. 1987 Jun;95(3):167–173. doi: 10.1111/j.1699-0463.1987.tb03107.x. [DOI] [PubMed] [Google Scholar]
- Olsvik O., Wasteson Y., Lund A., Hornes E. Pathogenic Escherichia coli found in food. Int J Food Microbiol. 1991 Jan;12(1):103–113. doi: 10.1016/0168-1605(91)90051-p. [DOI] [PubMed] [Google Scholar]
- Perry R. D., Brubaker R. R. Vwa+ phenotype of Yersinia enterocolitica. Infect Immun. 1983 Apr;40(1):166–171. doi: 10.1128/iai.40.1.166-171.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy D. A., Martinez R. J. Role of a plasmid in the pathogenicity of Yersinia species. Curr Top Microbiol Immunol. 1985;118:29–51. doi: 10.1007/978-3-642-70586-1_3. [DOI] [PubMed] [Google Scholar]
- Skjerve E., Rørvik L. M., Olsvik O. Detection of Listeria monocytogenes in foods by immunomagnetic separation. Appl Environ Microbiol. 1990 Nov;56(11):3478–3481. doi: 10.1128/aem.56.11.3478-3481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik M., Wolf-Watz H. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol Microbiol. 1989 Apr;3(4):517–529. doi: 10.1111/j.1365-2958.1989.tb00198.x. [DOI] [PubMed] [Google Scholar]
- Viitanen A. M., Arstila T. P., Lahesmaa R., Granfors K., Skurnik M., Toivanen P. Application of the polymerase chain reaction and immunofluorescence techniques to the detection of bacteria in Yersinia-triggered reactive arthritis. Arthritis Rheum. 1991 Jan;34(1):89–96. doi: 10.1002/art.1780340114. [DOI] [PubMed] [Google Scholar]
- Wahlberg J., Lundeberg J., Hultman T., Uhlén M. General colorimetric method for DNA diagnostics allowing direct solid-phase genomic sequencing of the positive samples. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6569–6573. doi: 10.1073/pnas.87.17.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauters G., Kandolo K., Janssens M. Revised biogrouping scheme of Yersinia enterocolitica. Contrib Microbiol Immunol. 1987;9:14–21. [PubMed] [Google Scholar]
- Wren B. W., Tabaqchali S. Detection of pathogenic Yersinia enterocolitica by the polymerase chain reaction. Lancet. 1990 Sep 15;336(8716):693–693. doi: 10.1016/0140-6736(90)92191-j. [DOI] [PubMed] [Google Scholar]