Abstract
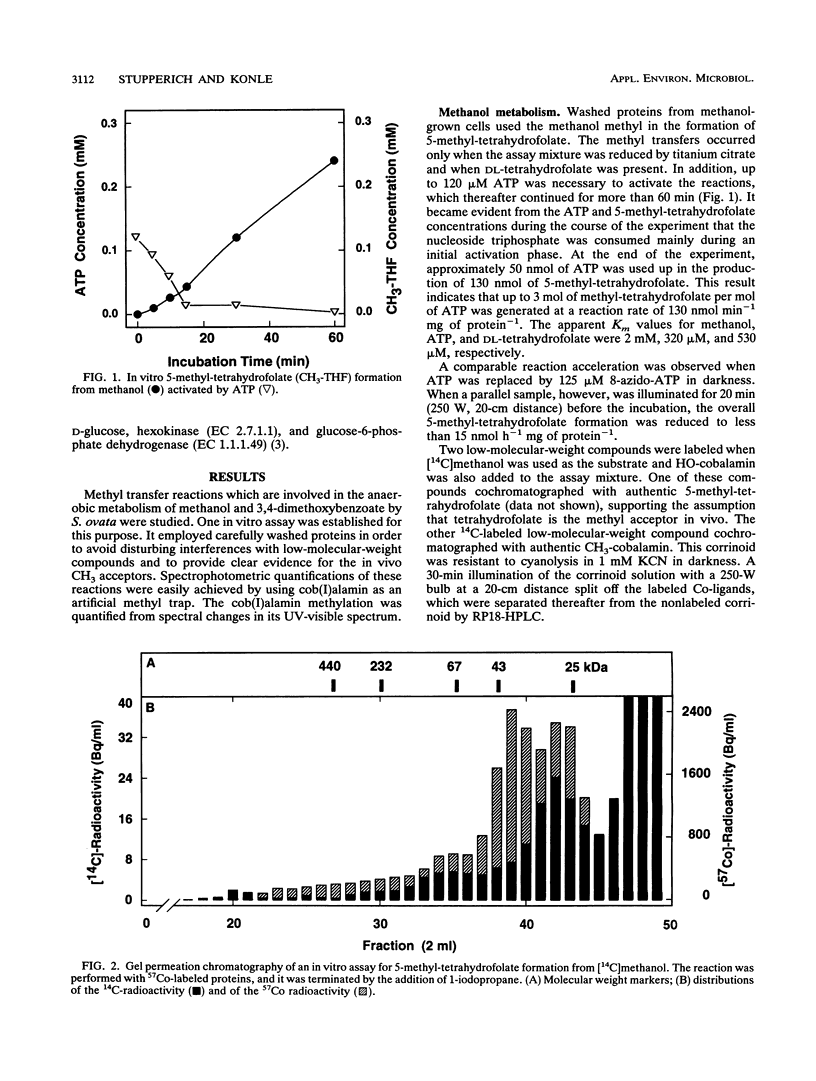
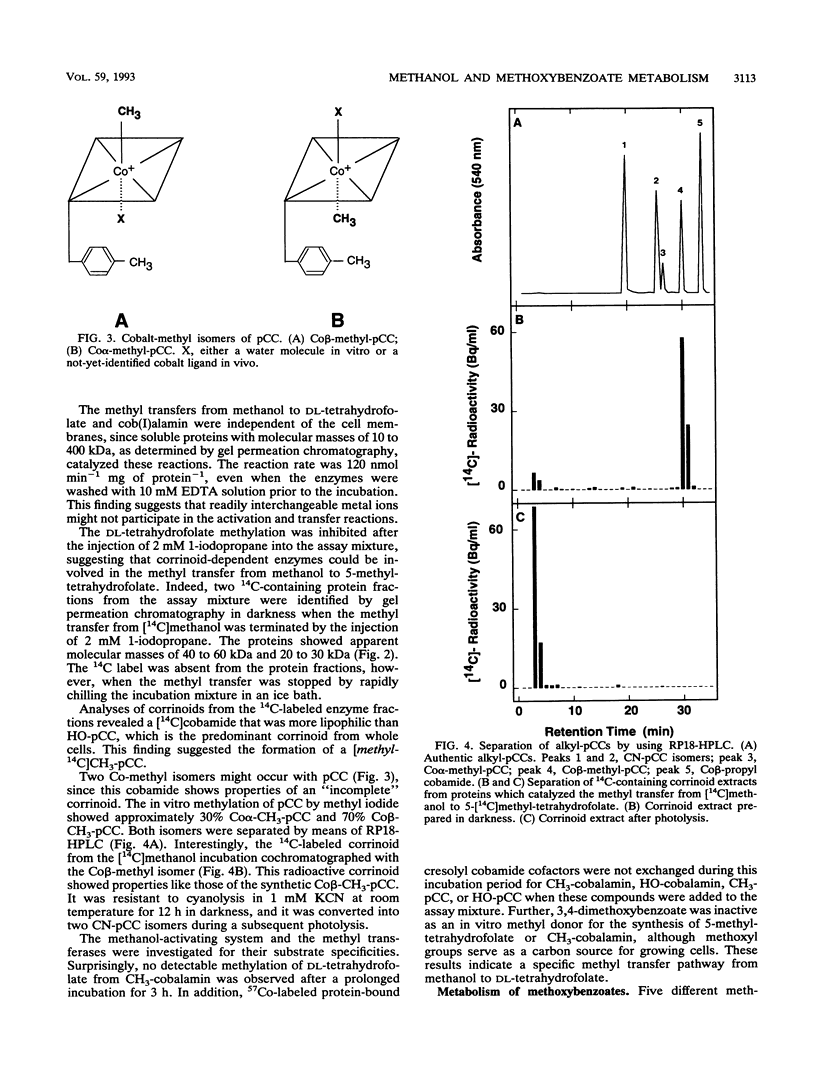
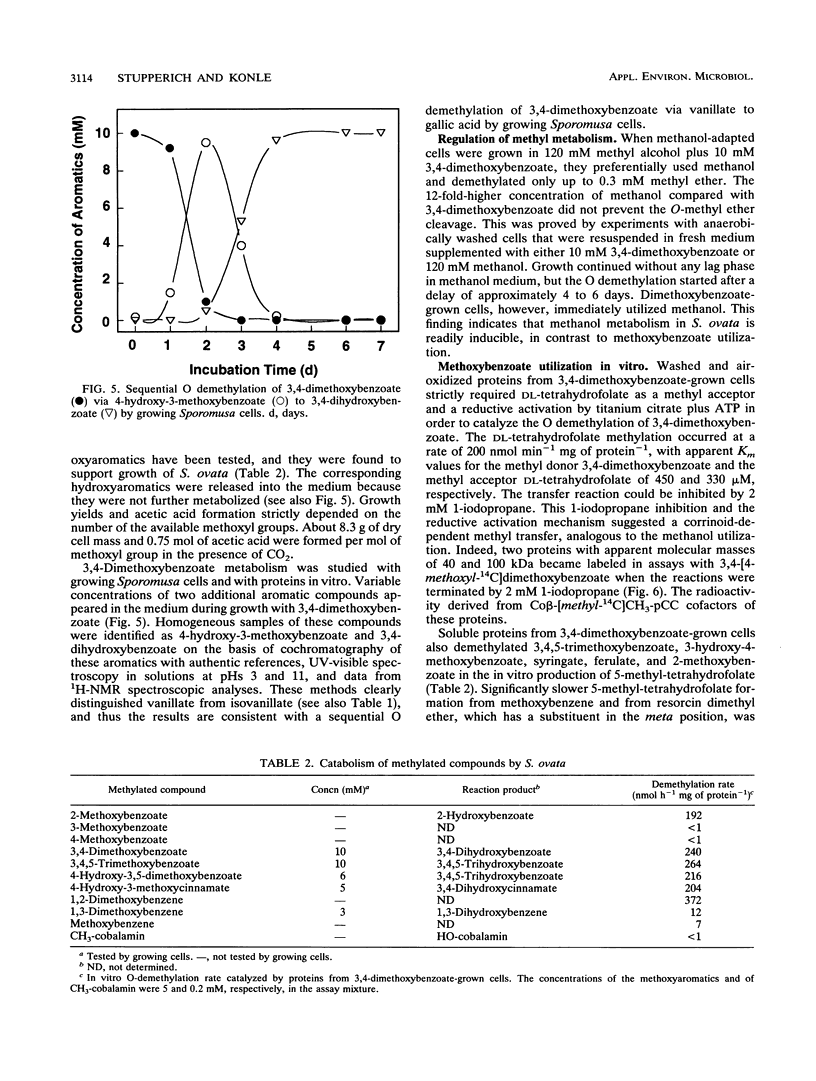
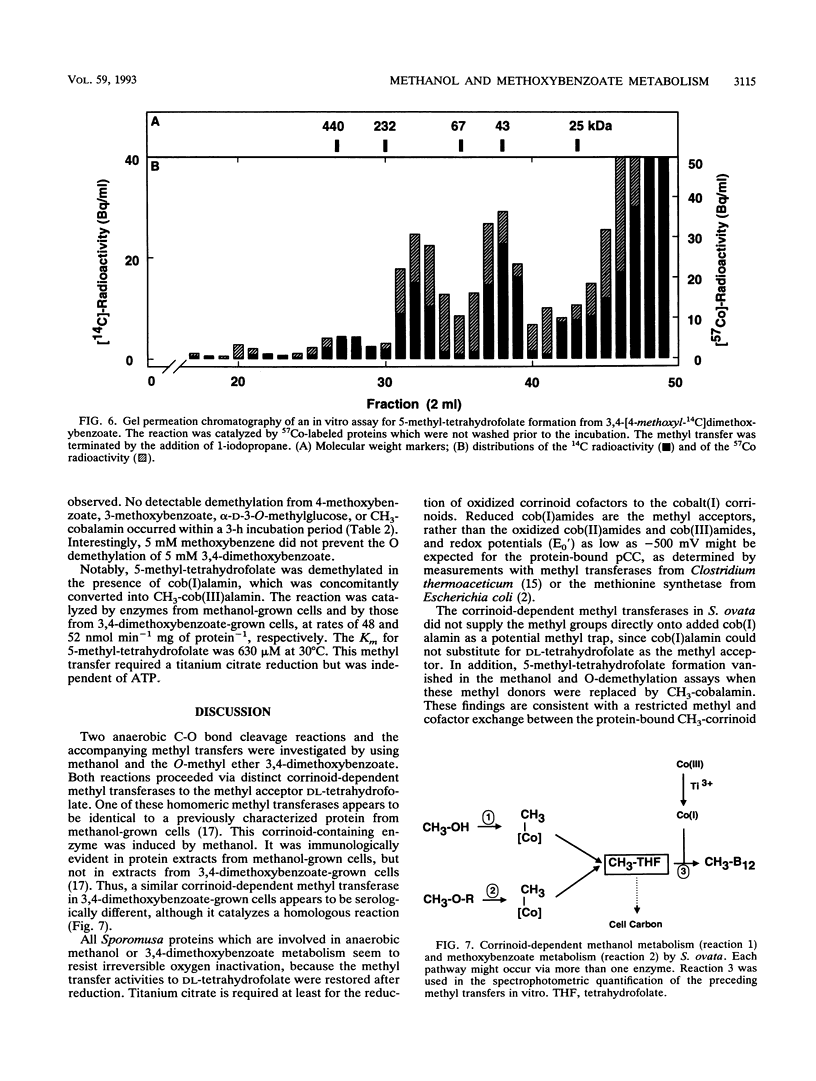
Washed and air-oxidized proteins from Sporomusa ovata cleaved the C-O bond of methanol or methoxyaromatics and transferred the methyl to dl-tetrahydrofolate. The reactions strictly required a reductive activation by titanium citrate, catalytic amounts of ATP, and the addition of dl-tetrahydrofolate. Methylcorrinoid-containing proteins carried the methanol methyl, which was transferred to dl-tetrahydrofolate at a specific rate of 120 nmol h-1 mg of protein-1. Tetrahydrofolate methylation diminished after the addition of 1-iodopropane or when the methyl donor methanol was replaced by 3,4-dimethoxybenzoate. However, whole Sporomusa cells utilize the methoxyl groups of 3,4-dimethoxybenzoate as a carbon source by a sequential O demethylation to 4-hydroxy-3-methoxybenzoate and 3,4-dihydroxybenzoate. The in vitro O demethylation of 3,4-[4-methoxyl-14C]dimethoxybenzoate proceeded via two distinct corrinoid-containing proteins to form 5-[14C]methyltetrahydrofolate at a specific rate of 200 nmol h-1 mg of protein-1. Proteins from 3,4-dimethoxybenzoate-grown cells efficiently used methoxybenzoates with vicinal substituents only, but they were unable to activate methanol. These results emphasized that specific enzymes are involved in methanol activation as well as in the activation of various methoxybenzoates and that similar corrinoid-dependent methyl transfer pathways are employed in 5-methyl-tetrahydrofolate formation from these substrates. Methyl-tetrahydrofolate could be demethylated by a distinct methyl transferase. That enzyme activity was present in washed and air-oxidized cell extracts from methanol-grown cells and from 3,4-dimethoxybenzoate-grown cells. It used cob(I)alamin as the methyl acceptor in vitro, which was methylated at a rate of 48 nmol min-1 mg of protein-1 even when ATP was omitted from the assay mixture. This methyl-cob(III)alamin formation made possible a spectrophotometric quantification of the preceding methyl transfers from methanol or methoxybenzoates to dl-tetrahydrofolate.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerjee R. V., Harder S. R., Ragsdale S. W., Matthews R. G. Mechanism of reductive activation of cobalamin-dependent methionine synthase: an electron paramagnetic resonance spectroelectrochemical study. Biochemistry. 1990 Feb 6;29(5):1129–1135. doi: 10.1021/bi00457a005. [DOI] [PubMed] [Google Scholar]
- Berman M. H., Frazer A. C. Importance of tetrahydrofolate and ATP in the anaerobic O-demethylation reaction for phenylmethylethers. Appl Environ Microbiol. 1992 Mar;58(3):925–931. doi: 10.1128/aem.58.3.925-931.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWeerd K. A., Saxena A., Nagle D. P., Jr, Suflita J. M. Metabolism of the 18O-methoxy substituent of 3-methoxybenzoic acid and other unlabeled methoxybenzoic acids by anaerobic bacteria. Appl Environ Microbiol. 1988 May;54(5):1237–1242. doi: 10.1128/aem.54.5.1237-1242.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer D. F., Tiedje J. M. Metabolism of polyethylene glycol by two anaerobic bacteria, Desulfovibrio desulfuricans and a Bacteroides sp. Appl Environ Microbiol. 1986 Oct;52(4):852–856. doi: 10.1128/aem.52.4.852-856.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer A. C., Young L. Y. A gram-negative anaerobic bacterium that utilizes o-methyl substituents of aromatic acids. Appl Environ Microbiol. 1985 May;49(5):1345–1347. doi: 10.1128/aem.49.5.1345-1347.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer A. C., Young L. Y. Anaerobic c(1) metabolism of the o-methyl-C-labeled substituent of vanillate. Appl Environ Microbiol. 1986 Jan;51(1):84–87. doi: 10.1128/aem.51.1.84-87.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić-Galić D. O-demethylation, dehydroxylation, ring-reduction and cleavage of aromatic substrates by Enterobacteriaceae under anaerobic conditions. J Appl Bacteriol. 1986 Dec;61(6):491–497. doi: 10.1111/j.1365-2672.1986.tb01721.x. [DOI] [PubMed] [Google Scholar]
- Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978 Jun 15;87(1):206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Ragsdale S. W. Enzymology of the acetyl-CoA pathway of CO2 fixation. Crit Rev Biochem Mol Biol. 1991;26(3-4):261–300. doi: 10.3109/10409239109114070. [DOI] [PubMed] [Google Scholar]
- Stupperich E., Aulkemeyer P., Eckerskorn C. Purification and characterization of a methanol-induced cobamide-containing protein from Sporomusa ovata. Arch Microbiol. 1992;158(5):370–373. doi: 10.1007/BF00245367. [DOI] [PubMed] [Google Scholar]
- Stupperich E., Eisinger H. J., Kräutler B. Identification of phenolyl cobamide from the homoacetogenic bacterium Sporomusa ovata. Eur J Biochem. 1989 Dec 22;186(3):657–661. doi: 10.1111/j.1432-1033.1989.tb15256.x. [DOI] [PubMed] [Google Scholar]
- Taylor B. F. Aerobic and Anaerobic Catabolism of Vanillic Acid and Some Other Methoxy-Aromatic Compounds by Pseudomonas sp. Strain PN-1. Appl Environ Microbiol. 1983 Dec;46(6):1286–1292. doi: 10.1128/aem.46.6.1286-1292.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. R., Daniel S. L., Drake H. L. Characterization of a CO-dependent O-demethylating enzyme system from the acetogen Clostridium thermoaceticum. J Bacteriol. 1988 Dec;170(12):5747–5750. doi: 10.1128/jb.170.12.5747-5750.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden P., te Brömmelstroet B. W., Poirot C. M., van der Drift C., Vogels G. D. Purification and properties of methanol:5-hydroxybenzimidazolylcobamide methyltransferase from Methanosarcina barkeri. J Bacteriol. 1984 Nov;160(2):629–635. doi: 10.1128/jb.160.2.629-635.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]


