Abstract
Most clock genes encode transcription factors that interact to elicit cooperative control of clock function. Using a two-hybrid system approach, we have isolated two different partners of zebrafish (zf) CLOCK, which are similar to the mammalian BMAL1 (brain and muscle arylhydrocarbon receptor nuclear translocator-like protein 1). The two homologs, zfBMAL1 and zfBMAL2, contain conserved basic helix–loop–helix-PAS (Period-Arylhydrocarbon receptor-Singleminded) domains but diverge in the carboxyl termini, thus bearing different transcriptional activation potential. As for zfClock, the expression of both zfBmals oscillates in most tissues in the animal. However, in many tissues, the peak, levels, and kinetics of expression are different between the two genes and for the same gene from tissue to tissue. These results support the existence of independent peripheral oscillators and suggest that zfBMAL1 and zfBMAL2 may exert distinct circadian functions, interacting differentially with zfCLOCK at various times in different tissues. Our findings also indicate that multiple controls may be exerted by the central clock and/or that peripheral oscillators can differentially interpret central clock signals.
The past few years have been characterized by a remarkable advance in our understanding of the molecular mechanisms underlying the biological clock, particularly in the identification of genes involved in clock function (1, 2). The Drosophila proteins Period (Per) and Timeless (Tim), and the product of Neurospora gene frequency (frq), have been known as clock components for some time (1, 2). The Clock gene was isolated by a genetic screen in mice (3) followed by positional cloning (4) and is the only bona fide clock gene in mammals. Clock homologs subsequently have been isolated in Drosophila (5) and zebrafish (6) by library screening at low stringency. In parallel, a Drosophila mutant in the gene encoding CLOCK was identified (7). In contrast to the mouse (8–10), Clock transcript levels exhibit a robust circadian oscillation both in flies and zebrafish (6, 11), possibly suggesting different regulation of clock gene function in various systems.
A significant number of clock genes products contain PAS (Period, Arylhydrocarbon receptor, Singleminded) and basic helix–loop–helix (bHLH) domains, structures that preside over the dimerization and DNA-binding functions of these transcription factors (1, 2). In Drosophila, for instance, Tim was found to be a partner of Per by using a yeast two-hybrid screen with the PAS domain of the Per protein as bait (12). The question as to what protein partners CLOCK might possess was tackled by the same approach in the mouse, leading to the identification of the protein BMAL1 [brain and muscle arylhydrocarbon receptor nuclear translocator (ARNT)-like protein 1] (13), a product also isolated by searches of orphan PAS-containing proteins in the databases (14, 15). Thus the PAS domain could be considered as a recurring feature in clock molecules, as also suggested by the recent identification of white-collar1 and white-collar2 in Neurospora (16).
The BMAL1 protein has been shown to associate with CLOCK and the heterodimers to regulate the expression of genes containing E-box elements in their regulatory region (13, 17). The CLOCK-BMAL1 heterodimers appear to function as transcriptional activators and in particular to induce the expression of per and tim genes in a circadian feedback loop (12, 18).
The zebrafish constitutes an attractive model to study the biological clock in vertebrates (6, 19, 20). A recent analysis of zebrafish Clock (zfClock) has shown its rhythmic pattern of expression in both retina and pineal gland (6). Remarkably, circadian expression also is observed in peripheral tissues, even when these are placed ex vivo for a number of days in culture dishes (6), as well as in a zebrafish-derived cell line (20). Moreover, the circadian clock of these cells and ex vivo organs can be directly entrained by light (20). These observations indicated the existence of independent circadian oscillators in peripheral tissues of the zebrafish.
The striking differences in the pattern of Clock expression between mouse and zebrafish hinted at possible diverse modes of regulation and function. We have been interested in identifying the CLOCK partners in zebrafish and studying their expression. The present study reports the isolation, by use of a two-hybrid screen, of two zebrafish cDNAs encoding distinct homologs of mammalian BMAL1. The analysis of the sequence, the expression pattern, the transcriptional activity, and the strength of association with CLOCK of these two zfBMALs highlights clear differences between them, indicating a way by which a fine-tuning of circadian gene expression could be achieved. Moreover, we report intertissue differences in the expression of each individual Bmal gene, supporting the existence of independent circadian oscillators in various tissues of the fish. Our results also indicate that multiple controls may be exerted by the central clock on peripheral oscillators and/or that peripheral oscillators are able to differentially interpret central clock signals.
Materials and Methods
Two-Hybrid System.
The portion of zfClock cDNA encoding amino acids 2–453 was amplified by PCR and cloned in the vector pGBT9 (CLONTECH), in-frame with GAL4 DNA-binding domain (DBD). This bait plasmid was cotransformed in CG-1945 yeast cells with a 1-month-old whole zebrafish cDNA library fused to GAL4 activation domain (AD) (CLONTECH, QL4000AB). Yeast two-hybrid screening was performed as described (CLONTECH Matchmaker Two-Hybrid System Protocol), with 40 mM 3-aminotriazole in the selection medium. β-Galactosidase assay was performed in Y190 yeast cells following the directions of the manufacturer. The results reported are in Miller units and are the means of assays performed in triplicate. For subsequent two-hybrid assays, complete or partial BMAL reading frames were cloned into pGAD424 (CLONTECH). Controls with pGBT9 (GAL4 DBD) or pLAM5′ (GAL4 DBD-lamin C) were always negative in β-galactosidase assays.
Sequence Analysis.
Sequence analysis and alignments were performed by using the University of Wisconsin GCG computer package, version 8.1. Phylogenetic trees were derived from a multiple alignment by using the phylip package (40), with 100 bootstrap resamplings. Mouse ARNT, ARNT2, and CLOCK were included as an outgroup to root the BMAL part of the tree. Only the unambiguously aligned portion of the sequences was kept to derive the phylogeny, which corresponds approximately to the bHLH-PAS region of the proteins. Similar results were obtained by using Fitch (Fig. 1D), neighbor-joining, or parsimony methods. The GenBank accession numbers of sequences used are: D89722 (human BMAL1a), AB000812 (human BMAL1b), AB014494 (mouse BMAL1 or Arnt3), AF065473 (Drosophila CYCLE), U14333 (mARNT), D63644 (mARNT2), and AF000998 (mCLOCK).
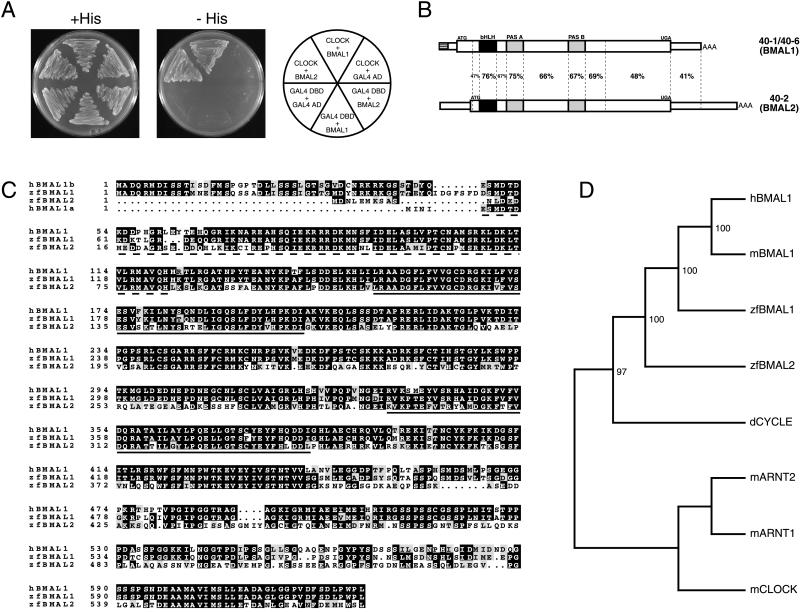
Figure 1.
Cloning and sequence analysis of zfBmal cDNAs. (A) Yeast growth is supported on selective medium in two-hybrid assays including both CLOCK and either of the zfBMALs (clones 40–1 and 40–2). (B) DNA sequence identity between the Bmal cDNAs. The hatched bar in 40–1/6 represents the region differing between the two clones. The ORF is denoted by a thicker box. (C) Alignment of human and zebrafish BMALs. Except for their amino terminus, human isoforms a and b are identical. Identical (inverted) and similar (gray background) residues are indicated. Hatched and solid underlined sequences highlight the bHLH domain and the PAS domain repeats, respectively. (D) Phylogenetic analysis of BMAL proteins. The result of 100 bootstrap resamplings is shown. mARNT1, mARNT2, and mCLOCK were used to root the tree (see Materials and Methods for details).
Glutathione S-Transferase (GST) Pull-Down Assay.
The bHLH-PAS region of the zfClock cDNA (as in the two-hybrid system) was amplified and cloned in pGEX4T-3 (Amersham Pharmacia). Extracts containing GST-CLOCK were prepared as in ref. 21, and the presence of the fusion protein was confirmed by Western analysis. The insert of two-hybrid clones 40–6 and 40–2 was amplified (the upstream primer containing a T7 promoter), and 35S-labeled BMAL1 and BMAL2 proteins were obtained by in vitro translation (TNT T7 Quick Coupled Transcription/Translation System, Promega). GST-CLOCK (or GST) was bound to glutathione Sepharose 4B beads (Amersham Pharmacia), and the GST pull-down with BMAL proteins was essentially as described in ref. 21.
RNA Analysis and in Situ Hybridization.
Zebrafish were grown and dissected as described (6). All fish were maintained on a 14:10 light-dark (LD) cycle or in constant darkness. RNA extractions were done as described (22) or by using TRIzol reagent (GIBCO/BRL). RNase protection assays (RPAs) were performed as described (6, 23). The Bmal riboprobes (from nucleotides 1,328–1,759 of Bmal1 reading frame and nucleotides 1,199–1,599 of Bmal2 reading frame, in pBluescript SK−) were generated by using an in vitro transcription kit (Promega). The probes extended over the most divergent part between Bmal1 and Bmal2 sequences, thereby allowing distinction between them in the assay. The experiments were repeated three times with similar results.
In situ hybridization on 10-μm serial cross sections of OCT-embedded fish brains was done as described (24), by using 35S-ATPαS-labeled probes prepared from the same plasmids as for the RPAs.
Results
Cloning and Sequence Analysis of Two zfBmal1 Homologs.
As an approach to identifying CLOCK partners in zebrafish a two-hybrid screen was performed. We used the bHLH-PAS moiety of zfCLOCK as a bait and a whole-zebrafish cDNA library fused to the GAL4 AD. Three clones of seven million were positive for both HIS3 and lacZ reporter gene expression (Figs. 1A and 2A). Clones 40–1 and 40–6 encoded the same protein, only the beginning of the 5′ untranslated region was different, possibly the result of alternative splicing. The third positive clone (40–2) encoded a different but related protein (Fig. 1B). The two proteins, called zfBMAL1 and zfBMAL2, share similarity with the mammalian protein BMAL1 (14, 15, 25) (Fig. 1C) and present an overall 59% identity and 75% similarity at the amino acid level between each other, with the C termini being most divergent. A phylogenetic tree of BMALs and other bHLH-PAS proteins reveals that zfBMAL1 is highly similar to human and rodent BMAL1, whereas zfBMAL2 is more divergent (Fig. 1D). The position within the tree of the Drosophila homolog of BMAL1, dCYCLE (26), suggests that the duplication that led to the two zebrafish genes occurred after the separation of the vertebrate and insect lineages. The zebrafish constitutes the first species where more than one Bmal gene is found.
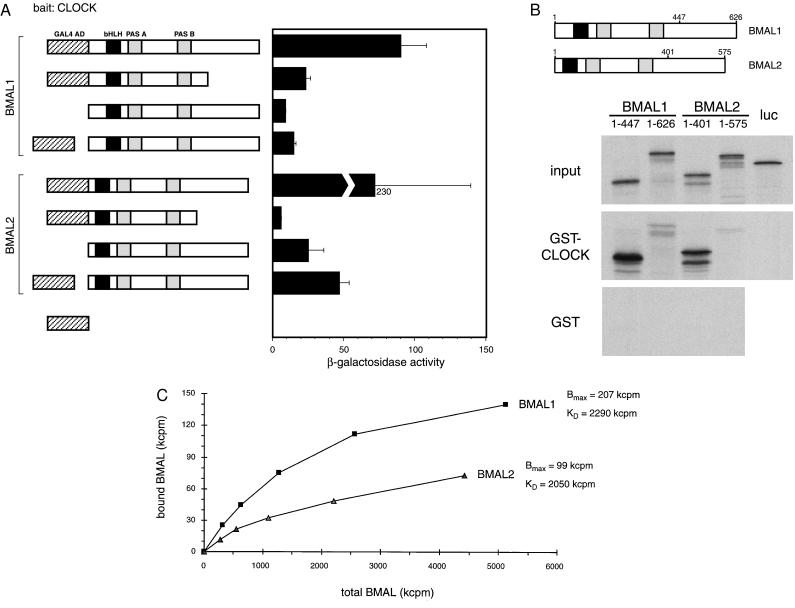
Figure 2.
Interaction of BMALs with CLOCK. (A) Yeast two-hybrid assays using various constructions in conjunction with GAL4 DBD-CLOCK. (Left) Representation of the different BMAL-based constructions. Hatched box denotes the GAL4 AD. (Right) β-Galactosidase activity, expressed in standard Miller units. The BMAL1 and BMAL2 constructions shown with a separated GAL4 AD (fourth and eighth from the top) correspond to the original clones isolated in the screen (40–1 and 40–2). (B) GST pull-down assay. Equivalent amounts of 35S-labeled complete or partial BMAL1 and BMAL2 proteins were prepared and analyzed by SDS/PAGE (input). (Lower) SDS/PAGE analysis of the proteins binding to GST or GST-CLOCK beads. Luciferase was used as a input negative control. (C) Different association affinities of zfBMAL proteins with CLOCK. 35S-labeled BMAL1 and BMAL2 were produced by in vitro transcription/translation, and radioactivity was quantified by trichloroacetic acid precipitation and scintillation counting. GST pull-down was carried out as described in Materials and Methods, with equivalent amounts of GST-CLOCK and increasing amounts of BMALs. After washes, the radioactivity remaining on the resin was quantified by scintillation counting.
Binding of the zfBMALs to zfCLOCK.
The significant sequence divergence within the two Bmal genes begged the question on possible differences in the functional properties of the relative protein products. The association of zfBMAL1 and zfBMAL2 with CLOCK was investigated in a two-hybrid system (Fig. 2A). When the bHLH-PAS region is fused to the GAL4 AD, activation of the reporter lacZ gene is observed, implying that this region is sufficient to promote heterodimerization of BMALs with CLOCK. Interestingly, BMAL1 interacts with CLOCK more efficiently than BMAL2 (Fig. 2A), whereas we have found no interaction between the two BMALs (not shown). Importantly, both BMALs have intrinsic activation potential as they strongly stimulate transcription even in the absence of the GAL4 AD. In a comparative study, BMAL2 acts as a much more potent transactivator than BMAL1.
The in vivo association of BMAL1 and BMAL2 with CLOCK observed in yeast was confirmed by a GST pull-down assay (Fig. 2B). Truncated proteins lacking the C terminus show that the bHLH-PAS region is sufficient to ensure association. Interestingly, the full-length proteins interact with reduced efficiency as compared with the truncated proteins, suggesting that the carboxyl terminus may in some way influence association. Also in this GST pull-down assay BMAL1 appears to associate more efficiently with CLOCK than BMAL2 (Fig. 2 B and C), confirming the results obtained in the two-hybrid assay. Thus, noteworthy differences exists between the two zfBMALs with respect to their regulatory capacity.
Expression of the zfBmal Genes in the Brain, Eyes, and Pineal Gland.
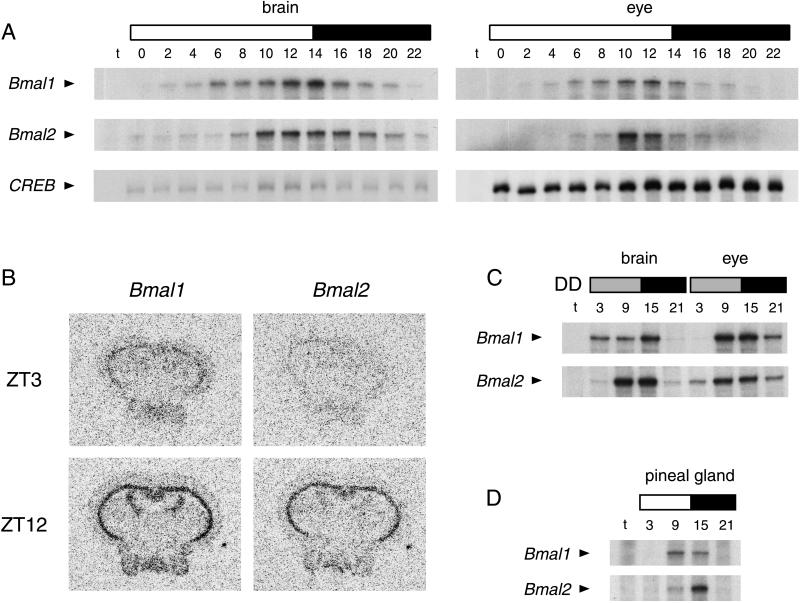
We previously have reported robust oscillation of Clock expression in clock structures, such as the eye and the pineal gland, but also in most peripheral tissues (6). This situation radically differs from what is known in mammals where Clock expression does not oscillate (8–10). This finding prompted us to analyze the expression of the two Bmals during the circadian cycle. In the brain both genes exhibit a robust rhythmic expression under LD conditions (Fig. 3A). The peak, however, is different for the two genes, Bmal1 reaching maximal expression at Zeitgeber time (ZT) 14, whereas Bmal2 peaks at ZT12. In addition, the kinetics of expression of the two genes differ, with the induction of Bmal2 being delayed but more rapid. The situation in the eye is similar, although the peak of Bmal1 expression is at ZT10–12 instead of ZT14 as in the brain (Fig. 3A). In situ hybridization analysis with zebrafish brains revealed that the distribution of the two Bmal transcripts is very similar (Fig. 3B) and that it overlaps that of Clock (6). Bmals are expressed at high levels in the periventricular gray zone of the optic tectum, valvula cerebelli, corpus mammilare, and hypothalamus. Importantly, robust rhythmic expression of both genes is maintained when the animals are kept in a dark-dark regime, both in the brain and eye (Fig. 3C). The other clock structure, the pineal gland, also shows significant rhythmic expression of both genes (Fig. 3D), but Bmal1 expression appears to peak earlier than for Bmal2. When compared with the rhythmic pattern of Clock expression (6) (see Table 1), it is evident that expression of the three genes is not synchronized in the brain and clock structures.
Figure 3.
Rhythmic expression of the Bmal genes in the zebrafish brain, eyes, and pineal gland. (A, C, and D) RPA of the expression of the Bmal transcripts in the brain, eyes, and pineal gland under LD (A and D) or constant darkness (C) conditions. Equivalent amounts of RNA were used in each case, as assayed by ethidium staining and CREB (cAMP responsive binding protein) RPA control. “t” is the negative control with only tRNA. Numbers above each lane correspond to the ZT points at which RNA samples were prepared. (A and D) White/black bars represent the duration of the LD periods. (C) Gray/black bars show the extent of subjective day/night periods in dark-dark (DD) conditions. (B) In situ hybridization of zebrafish brain sections at ZT3 or ZT12 (in LD). For each time point, two successive sections were analyzed with Bmal1 and Bmal2 probes.
Table 1.
Expression peaks of BmaI and Clock genes
| BmaI1 | BmaI2 | Clock* | |
|---|---|---|---|
| Brain† | ZT14 | ZT10 | ZT14–16 |
| Eye‡ | ZT10–12 | ZT10 | ZT12 |
| Heart | ∼ZT9 | ∼ZT9 | ∼ZT15 |
| Kidney | ∼ZT15 | ∼ZT9–15 | ∼ZT15 |
| Liver | ∼ZT9 | ∼ZT15 | ∼ZT9–15 |
| Pineal gland | ∼ZT9–15 | ∼ZT15 | ∼ZT15 |
| Spleen | ∼ZT15 | ∼ZT15 | ∼ZT15 |
| Testis | No oscillation | No oscillation | No oscillation |
Data published in ref. 6, except for the liver (N.C., unpublished results).
† Different kinetics of accumulation for BmaI1 and BmaI2.
‡ Different kinetics of accumulation for BmaI1 and BmaI2; Clock kinetics are intermediate.
Expression of the zfBmal Genes in Other Tissues.
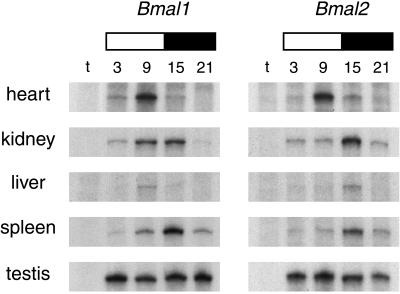
We recently have shown that, in contrast to mammals (8–10), Clock expression in the zebrafish strongly oscillates in various tissues (6). Importantly Clock rhythmicity is synchronous throughout the animal (6), which indicated that a common clock control is likely to be exerted on peripheral tissues. Analysis of Bmal1 and Bmal2 expression in various tissues revealed a strikingly discordant scenario (Fig. 4). Besides testis, where as for Clock no oscillation is detected, robust rhythmicity is observed in all tissues. Remarkably, oscillations have different phases for each gene in various tissues and for the two genes in the same tissue. When compared with Clock, it is evident that the oscillations displayed by the three partners are discordant (Table 1).
Figure 4.
Differential expression patterns of Bmal1 and Bmal2 in different zebrafish peripheral organs. RPA analysis of Bmal expression in adult zebrafish heart, kidney, liver, spleen, and testis at the indicated ZT points under LD conditions. “t” corresponds to a tRNA negative control, and white/black bars represent the day/night periods.
Discussion
We report here the isolation and characterization of two partners of the zfCLOCK protein, which are homologous to the mammalian BMAL1 protein. We show that the two proteins differ in their ability to associate with CLOCK and in their transcription activation potential. Their expression in various tissues displays robust oscillatory patterns, which differ from gene to gene in the same tissue and from tissue to tissue for the same gene.
Our results indicate that the molecular gears, and possibly the function, of the circadian clock are not conserved among vertebrates (28). In zebrafish there are two Bmal genes, whereas mammals have only one (14, 15). Although Clock expression is constant in mammals (8–10), it is oscillating in the fish (6). Finally, Bmal1 expression oscillates synchronously in most tissues of the rat (10), whereas here we show that this is not the case in the zebrafish (Fig. 4). It is interesting to note that expression of this gene in rodents and fish is almost antiphase; it is not clear, however, if this is because of profound differences in the organization of the molecular clock or whether it reflects the fact that rodents are nocturnal whereas fish are diurnal.
The presence of two Bmal genes in zebrafish, and their differential mRNA expression, suggests dynamic combinatorial associations with CLOCK in various tissues and at different times of the circadian cycle, although the protein levels may change with different phases or amplitudes. The different association potential of the two BMAL proteins with CLOCK and their diverse transactivation capacities (Fig. 2) indicate that resulting complexes might act on various target genes depending on the time of the circadian cycle. The picture might even be more complex: in some tissues (e.g., in the heart) both Bmals expression peaks when Clock is at very low levels (Table 1 and ref. 6), suggesting that BMALs may have other partners in addition to CLOCK. This would be consistent with the fact that mammalian BMAL1 is able to interact with other bHLH-PAS factors besides CLOCK (17, 25). The observation that the two BMALs exhibit different strength of transactivation and association with CLOCK suggests an additional layer of complexity in the regulation of gene expression by these bHLH-PAS factor heterodimers. This adds to the constraints imposed by temporal and tissue-specific expression of these factors.
Recent results in various systems point to the existence of local clocks in peripheral tissues (6, 20, 27–32). Several reports on circadian gene expression in rodents indicate that oscillations in the central clock structure, the suprachiasmatic nuclei, precede by several hours oscillations in peripheral tissues (8, 31, 33–36). This observation would infer the existence of ancillary clocks, located in peripheral tissues, which coordinately and synchronously respond to a central clock control (37, 38). We show that this possible scenario does not apply to all vertebrates. Indeed, in zfBmals peaks of expression in peripheral tissues may even precede the peak in the eye and the pineal gland (Figs. 3 and 4). This notion indicates that the transcriptional machinery controlling a given clock gene must be able to respond differentially in various tissues, to ensure oscillation with variable phase and period. It is possible that the central clock may be able to exert multiple controls through different signals to obtain distinct responses in each peripheral tissue (39). Another nonexclusive possibility is that peripheral oscillators may differentially interpret central clock signals.
Acknowledgments
We thank Estelle Heitz and Maryam Rastegar for their expert technical assistance and Uwe Strähle and all of the members of the Sassone-Corsi's laboratory for help, reagents, and discussions. N.C. dedicates this work to the memory of Robert Cedergren. N.C. was supported by a Human Frontier Science Program Organization long-term fellowship. D.W. was supported by a European Community fellowship. This work was supported by grants from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Hospitalier Régional Universitaire, Fondation pour la Recherche Médicale, and Association pour la Recherche sur le Cancer.
Abbreviations
- ARNT
arylhydrocarbon receptor nuclear translocator
- BMAL
brain and muscle ARNT-like
- PAS
Period-Arylhydrocarbon receptor-Singleminded
- zf
zebrafish
- bHLH
basic helix–loop–helix
- AD
activation domain
- ZT
Zeitgeber time
- DBD
DNA-binding domain
- LD
light-dark
- Per
Period
- GST
glutathione S-transferase
- RPA
RNase protection assay
Footnotes
References
- 1.Sassone-Corsi P. Nature (London) 1998;362:871–874. doi: 10.1038/31821. [DOI] [PubMed] [Google Scholar]
- 2.Dunlap J C. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 3.Vitaterna M H, King D P, Chang A M, Kornhauser J M, Lowrey P L, McDonald J D, Dove W F, Pinto L H, Turek F W, Takahashi J S. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King D P, Zhao Y, Sangoram A M, Wilsbacher L D, Tanaka M, Antoch M P, Steeves T D, Vitaterna M H, Kornhauser J M, Lowrey P L, et al. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darlington T K, Wager-Smith K, Ceriani M F, Staknis D, Gekakis N, Steeves T D, Weitz C J, Takahashi J S, Kay S A. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 6.Whitmore D, Foulkes N S, Strähle U, Sassone-Corsi P. Nat Neurosci. 1998;1:701–707. doi: 10.1038/3703. [DOI] [PubMed] [Google Scholar]
- 7.Allada R, White N E, So W V, Hall J C, Rosbash M. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 8.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 9.Shearman L P, Zylka M J, Reppert S M, Weaver D R. Neuroscience. 1999;89:387–397. doi: 10.1016/s0306-4522(98)00325-x. [DOI] [PubMed] [Google Scholar]
- 10.Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N. Biochem Biophys Res Commun. 1998;253:199–203. doi: 10.1006/bbrc.1998.9779. [DOI] [PubMed] [Google Scholar]
- 11.Bae K, Lee C, Sidote D, Chuang K Y, Edery I. Mol Cell Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gekakis N, Saez L, Delahaye-Brown A M, Myers M P, Sehgal A, Young M W, Weitz C J. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 13.Gekakis N, Staknis D, Nguyen H B, Davis F C, Wilsbacher L D, King D P, Takahashi J S, Weitz C J. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 14.Hogenesch J B, Chan W K, Jackiw V H, Brown R C, Gu Y Z, Pray-Grant M, Perdew G H, Bradfield C A. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda M, Nomura M. Biochem Biophys Res Commun. 1997;233:258–264. doi: 10.1006/bbrc.1997.6371. [DOI] [PubMed] [Google Scholar]
- 16.Crosthwaite S K, Dunlap J C, Loros J J. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 17.Hogenesch J B, Gu Y Z, Jain S, Bradfield C A. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sangoram A M, Saez L, Antoch M P, Gekakis N, Staknis D, Whiteley A, Fruechte E M, Vitaterna M H, Shimomura K, King D P, et al. Neuron. 1998;21:1101–1113. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 19.Hurd M W, Debruyne J, Straume M, Cahill G M. Physiol Behav. 1998;65:465–472. doi: 10.1016/s0031-9384(98)00183-8. [DOI] [PubMed] [Google Scholar]
- 20.Whitmore D, Foulkes N S, Sassone-Corsi P. Nature (London) 2000;400:87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- 21.Fimia G M, De Cesare D, Sassone-Corsi P. Nature (London) 1999;398:165–169. doi: 10.1038/18237. [DOI] [PubMed] [Google Scholar]
- 22.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 23.Foulkes N S, Borrelli E, Sassone-Corsi P. Cell. 1991;64:739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 24.Mellström B, Naranjo J R, Foulkes N S, Lafarga M, Sassone-Corsi P. Neuron. 1993;10:655–665. doi: 10.1016/0896-6273(93)90167-p. [DOI] [PubMed] [Google Scholar]
- 25.Takahata S, Sogawa K, Kobayashi A, Ema M, Mimura J, Ozaki N, Fujii- Kuriyama Y. Biochem Biophys Res Commun. 1998;248:789–794. doi: 10.1006/bbrc.1998.9012. [DOI] [PubMed] [Google Scholar]
- 26.Rutila J E, Suri V, Le M, So W V, Rosbash M, Hall J C. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 27.Menaker M, Moreira L F, Tosini G. Braz J Med Biol Res. 1997;30:305–313. doi: 10.1590/s0100-879x1997000300003. [DOI] [PubMed] [Google Scholar]
- 28.Tosini G, Menaker M. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 29.Plautz J D, Kaneko M, Hell J C, Kay S A. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 30.Balsalobre A, Damiola F, Schibler U. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 31.Zylka M J, Shearman L P, Weaver D R, Reppert S M. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 32.Krishnan B, Dryer S E, Hardin P E. Nature (London) 1999;400:375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- 33.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Nature (London) 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 34.Shearman L P, Zylka M J, Weaver D R, Kolakowski L F, Reppert S M. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 35.Honma S, Ikeda M, Abe H, Tanahashi Y, Namihira M, Honma K, Nomura M. Biochem Biophys Res Commun. 1998;250:83–87. doi: 10.1006/bbrc.1998.9275. [DOI] [PubMed] [Google Scholar]
- 36.Abe H, Honma S, Namihira M, Tanahashi Y, Ikeda M, Honma K. Neurosci Lett. 1998;258:93–96. doi: 10.1016/s0304-3940(98)00877-5. [DOI] [PubMed] [Google Scholar]
- 37.Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, Sato K, Miyake Y, Ohara O, Kako K, Ishida N. J Biol Chem. 1998;273:27039–27042. doi: 10.1074/jbc.273.42.27039. [DOI] [PubMed] [Google Scholar]
- 38.Lucas R J, Stirland J A, Darrow J M, Menaker M, Loudon A S. Endocrinology. 1999;140:758–764. doi: 10.1210/endo.140.2.6538. [DOI] [PubMed] [Google Scholar]
- 39.Hastings M H. Trends Neurosci. 1997;20:459–464. doi: 10.1016/s0166-2236(97)01087-4. [DOI] [PubMed] [Google Scholar]
- 40.Felsenstein J. phylip, Phylogeny Inference Package, Version 3.5c. Seattle: Univ. of Washington; 1993. [Google Scholar]