Abstract
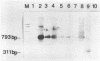
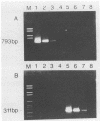
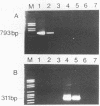
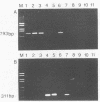
Antigen capture polymerase chain reaction (PCR) was tested as a sensitive and rapid method for detecting hepatitis A virus (HAV) in raw sewage sludge. The antigen capture PCR was performed both with and without solid-phase virus-catching monoclonal antibodies. Similar results proved that both methods were equally sensitive. Sewage sludge samples from different regions in Germany were examined for evidence of HAV contamination by antigen capture PCR. This method of detection was compared with that used in a previous study of these sewage sludge samples, in which the HAV was detected through indirect immunofluorescence after cell culture inoculation. The results obtained by antigen capture PCR matched those obtained in the earlier cell culture investigations, when HAV was detected in raw as well as digested sewage sludge samples. The advantage of the PCR method, however, lies in the fact that it needs only two days while the cell culture propagation of HAV takes about 8 to 10 weeks.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cohen J. I., Ticehurst J. R., Purcell R. H., Buckler-White A., Baroudy B. M. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J Virol. 1987 Jan;61(1):50–59. doi: 10.1128/jvi.61.1.50-59.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldadah Z. A., Asher D. M., Godec M. S., Pomeroy K. L., Goldfarb L. G., Feinstone S. M., Levitan H., Gibbs C. J., Jr, Gajdusek D. C. Detection of flaviviruses by reverse-transcriptase polymerase chain reaction. J Med Virol. 1991 Apr;33(4):260–267. doi: 10.1002/jmv.1890330410. [DOI] [PubMed] [Google Scholar]
- Flehmig B. Hepatitis A virus in cell culture. II. Growth characteristics of hepatitis A virus in Frhk-4/R cells. Med Microbiol Immunol. 1981;170(2):73–81. doi: 10.1007/BF02122671. [DOI] [PubMed] [Google Scholar]
- Gust I. D., Coulepis A. G., Feinstone S. M., Locarnini S. A., Moritsugu Y., Najera R., Siegl G. Taxonomic classification of hepatitis A virus. Intervirology. 1983;20(1):1–7. doi: 10.1159/000149367. [DOI] [PubMed] [Google Scholar]
- Jansen R. W., Siegl G., Lemon S. M. Molecular epidemiology of human hepatitis A virus defined by an antigen-capture polymerase chain reaction method. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2867–2871. doi: 10.1073/pnas.87.8.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Estes M. K., Metcalf T. G., Melnick J. L. Detection of hepatitis A virus in seeded estuarine samples by hybridization with cDNA probes. Appl Environ Microbiol. 1986 Oct;52(4):711–717. doi: 10.1128/aem.52.4.711-717.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Scholz E., Heinricy U., Flehmig B. Acid stability of hepatitis A virus. J Gen Virol. 1989 Sep;70(Pt 9):2481–2485. doi: 10.1099/0022-1317-70-9-2481. [DOI] [PubMed] [Google Scholar]
- Vallbracht A., Hofmann L., Wurster K. G., Flehmig B. Persistent infection of human fibroblasts by hepatitis A virus. J Gen Virol. 1984 Mar;65(Pt 3):609–615. doi: 10.1099/0022-1317-65-3-609. [DOI] [PubMed] [Google Scholar]