Abstract
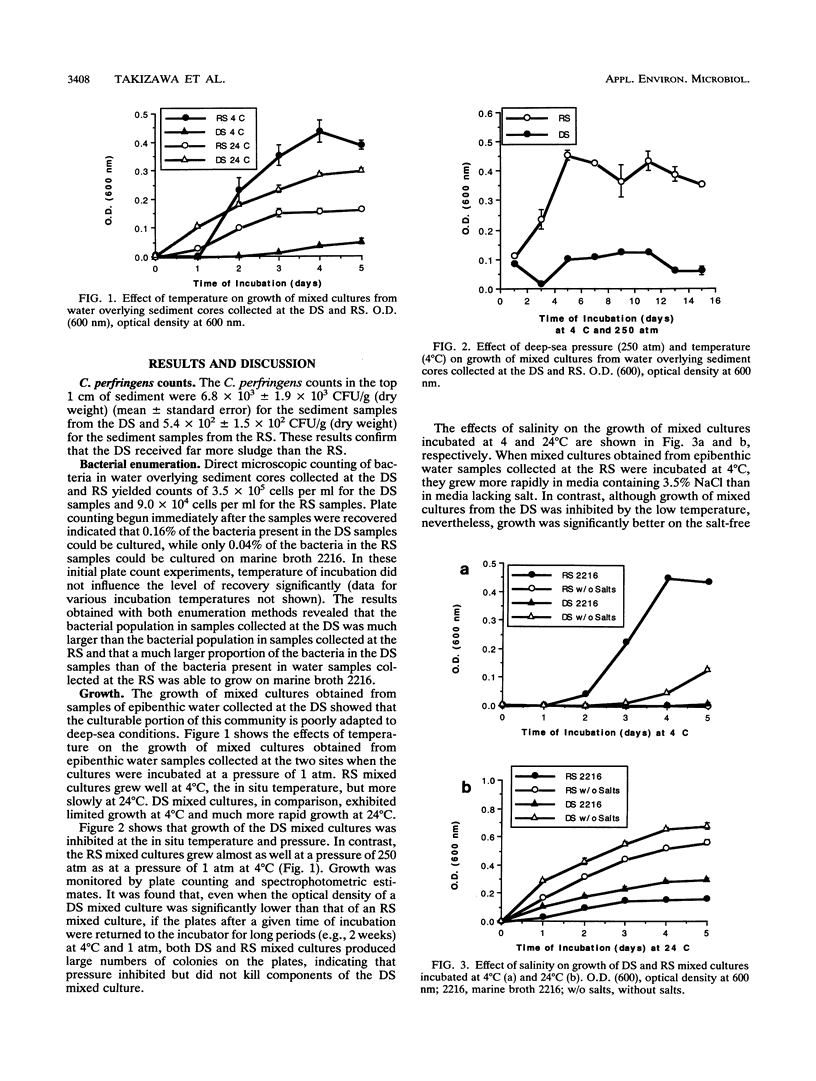
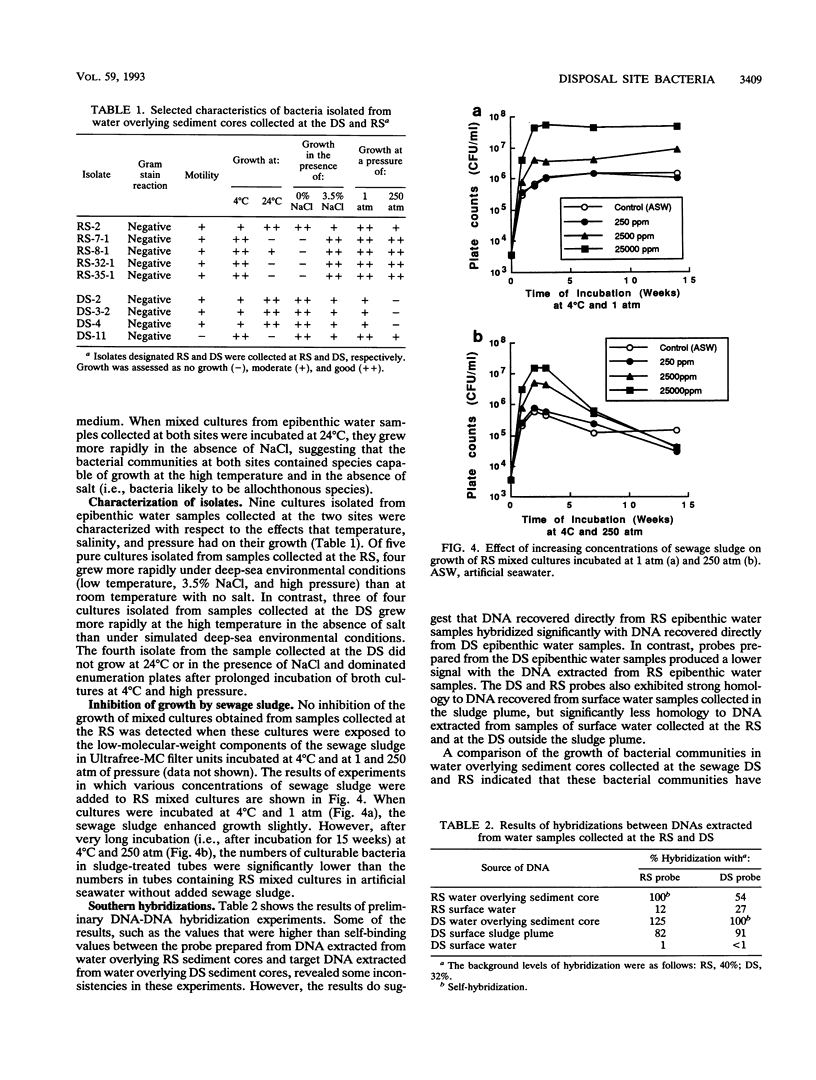
The epibenthic bacterial community at deep-ocean sewage sludge disposal site DWD-106, located approximately 106 miles (ca. 196 km) off the coast of New Jersey, was assessed for changes associated with the introduction of large amounts of sewage sludge. Mixed cultures and bacterial isolates obtained from water overlying sediment core samples collected at the deep-water (2,500 m) municipal sewage disposal site were tested for the ability to grow under in situ conditions of temperature and pressure. The responses of cultures collected at a DWD-106 station heavily impacted by sewage sludge were compared with those of samples collected from a station at the same depth which was not contaminated by sewage sludge. Significant differences were observed in the ability of mixed bacterial cultures and isolates from the two sites to grow under deep-sea pressure and temperature conditions. The levels of sludge contamination were established by enumerating Clostridium perfringens, a sewage indicator bacterium, in sediment samples from the two sites. The results of hybridization experiments in which DNAs extracted directly from the water overlying sediment core samples were used indicate that the reference site epibenthic community, the disposal site epibenthic community, and the community in a surface sludge plume share many members. Decreased culturability of reference site mixed cultures in the presence of sewage sludge was observed. Thus, the culturable portions of both the autochthonous and allochthonous bacterial communities at the disposal site may be inhibited in situ, the former by sewage sludge and the latter by high pressure and low temperature.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bisson J. W., Cabelli V. J. Membrane filter enumeration method for Clostridium perfringens. Appl Environ Microbiol. 1979 Jan;37(1):55–66. doi: 10.1128/aem.37.1.55-66.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R. T., Knight I. T., Anikis M. S., Colwell R. R. Benthic Distribution of Sewage Sludge Indicated by Clostridium perfringens at a Deep-Ocean Dump Site. Appl Environ Microbiol. 1993 Jan;59(1):47–51. doi: 10.1128/aem.59.1.47-51.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie J. E., Daley R. J., Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 May;33(5):1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Fuhrman J. A. DNA hybridization to compare species compositions of natural bacterioplankton assemblages. Appl Environ Microbiol. 1990 Mar;56(3):739–746. doi: 10.1128/aem.56.3.739-746.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K. L., Morita R. Y. Effects of hydrostatic pressure and temperature on the uptake and respiration of amino acids by a facultatively psychrophilic marine bacterium. J Bacteriol. 1971 Nov;108(2):835–843. doi: 10.1128/jb.108.2.835-843.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987 Sep;51(3):365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. C., Knight I. T., Straube W. L., Colwell R. R. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol. 1989 Mar;55(3):548–554. doi: 10.1128/aem.55.3.548-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yayanos A. A., Dietz A. S., Van Boxtel R. Dependence of reproduction rate on pressure as a hallmark of deep-sea bacteria. Appl Environ Microbiol. 1982 Dec;44(6):1356–1361. doi: 10.1128/aem.44.6.1356-1361.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


