Abstract
Members of the nuclear receptor superfamily are thought to activate transcription by recruitment of one or more recently identified coactivator complexes. Here we demonstrate that both peroxisome proliferator-activated receptor binding protein (PBP) and steroid receptor coactivator-1 (SRC-1) are required for ligand-dependent transcription of transiently transfected and chromosomally integrated reporter genes by the estrogen receptor (ER) and retinoic acid receptor (RAR). To examine ligand-dependent interactions between nuclear receptors and specific coactivators in living cells, these proteins were tagged with cyan (CFP) and yellow (YFP) mutants of the green fluorescent protein. Fluorescence resonance energy transfer (FRET) from the CFP to the YFP indicated interaction between the receptor and coactivator. CFP fusions to RAR or its ligand-binding domain exhibited rapid ligand-dependent FRET to YFP-tagged nuclear receptor interaction domains of the coactivators SRC-1 and PBP. The ER-ligand-binding domain, unlike RAR, also exhibited some basal interaction with coactivators in unstimulated cells that was abolished by the receptor antagonists tamoxifen or ICI182,780. Inhibition of FRET by tamoxifen but not ICI182,780 could be reversed by estradiol, whereas estradiol-enhanced FRET could not be inhibited by either antagonist, indicating that ligand effects can show varying degrees of hysteresis. These findings suggest that ligand-dependent transcriptional activities of the RAR and ER require concurrent or sequential recruitment of SRC-1 and PBP-containing coactivator complexes.
Nuclear receptors comprise a large family of ligand-dependent transcription factors that regulate diverse aspects of development and homeostasis (1). Members of this family contain a highly conserved DNA-binding domain that mediates sequence-specific interactions with response elements in target genes and a C-terminal ligand-binding domain (LBD) that mediates ligand-dependent transcriptional activation. Nuclear receptors that are regulated by steroid hormones, such as the estrogen receptor (ER), typically bind to their cognate response elements as homodimers, whereas a larger group of nuclear receptors, exemplified by retinoic acid receptors (RARs), interact with target genes as heterodimers with retinoid X receptors (2). Crystal structures of nuclear receptor LBDs in the presence and absence of ligands indicate that the binding of agonists induces a conformational change in a helical motif, referred to as AF-2, that allows nuclear receptors to couple to coactivator proteins that mediate their transcription effects (3). In contrast, binding of antagonists induces a conformational change in the AF-2 helix that prevents coactivator interaction (4, 5).
Members of a family of 160-kDa proteins, referred to as the steroid receptor coactivator (SRC) family, are among the best characterized nuclear receptor coactivators at the molecular and biochemical levels. Members of this family, which include SRC-1 and p300/cAMP responsive element binding (CREB)-interacting protein (p/CIP), interact with nuclear receptors through a central domain that contains three copies of a conserved recognition motif that contains the consensus sequence LXXLL (6–9). Each of these motifs adopts an α-helical structure that interacts with the nuclear receptor LBD in an AF-2 and ligand-dependent manner (5, 10, 11).
In addition to stimulating the ligand-dependent transcriptional activities of many nuclear receptors (12), microinjection of antibodies to specific members of the SRC family has been shown to block transactivation by RAR and ER (13, 14), suggesting that the SRC factors play essential roles in ligand-dependent activation in cells. The SRC factors are thought to function in part by recruiting the general coactivators CREB binding protein (CBP) and p300, which possess histone acetyltransferase activity and interact with other components of the transcriptional machinery (15).
Biochemical purification of factors associated with liganded thyroid hormone and vitamin D receptors have resulted in the identification of a multiprotein complex, referred to as the TRAP/DRIP (thyroid hormone receptor-associated protein/vitamin D receptor-interacting protein) complex, that stimulates their activities in in vitro transcription assays (16–18). This complex does not contain SRC or CBP/p300 factors and lacks histone acetyltransferase activity, suggesting that it serves a distinct transcriptional function. Interaction of the TRAP/DRIP complex with liganded nuclear receptors is mediated by a 220-kDa component equivalent to peroxisome proliferator-activated receptor binding protein (PBP; also known as TRAP220/DRIP220) (16, 19). This factor contains two LXXLL motifs and competes with SRC factors for interaction with liganded nuclear receptors; thus, two questions arise: do PBP and SRC-1 bind the same receptors within cells, and if they do, are they components of alternative transactivation pathways or are both required for receptor activation?
A new approach in the study of nuclear receptor signaling has been the use of chimeric fusions of the green fluorescent protein (GFP) with nuclear receptors. Fusions with GFP have been shown to be functional receptors, and the natural distribution of the receptors and changes in subnuclear staining in response to hormone have been reported (20). Fusions with GFP mutants have also been used to monitor protein–protein interaction by fluorescence resonance energy transfer (FRET) (refs. 21–23; see also Fig. 2A). We have now tagged the RAR and ER with cyan fluorescent protein (CFP), a cyan mutant of GFP, and the nuclear receptor interaction domains of SRC-1 and PBP with yellow fluorescent protein (YFP), a yellow mutant of GFP. Upon coexpression of appropriate pairs in the nucleus of HeLa cells, we observe changes in FRET consistent with protein association in response to hormone stimulation, and protein dissociation with receptor antagonists. In concert with microinjection experiments that reveal an essential role of PBP in ER and RAR function, the findings are consistent with a model of transcriptional activation requiring sequential or concurrent interactions of distinct coactivator complexes.
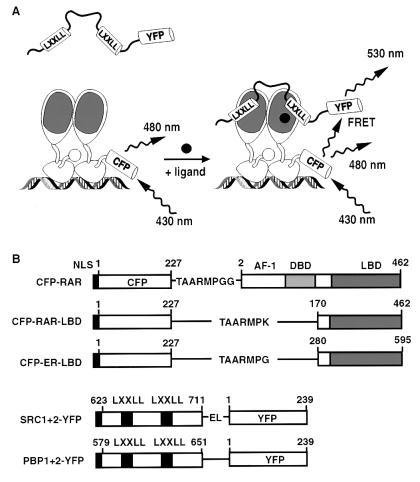
Figure 2.
FRET experimental design. (A) Ligand-dependent FRET illustrated for full-length CFP–RAR bound to DNA as a heterodimer with retinoid X receptors and YFP-tagged nuclear receptor interaction domain of SRC-1. (B) CFP and YFP fusion proteins used for FRET experiments. Junctional amino acids are indicated in single-letter code.
Materials and Methods
Materials.
All trans-retinoic acid (RA) and tamoxifen were from Sigma; (4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid (TTNPB) and 17β-estradiol (E2) were from Calbiochem; and ICI182,780 was from Tocris Cookson (Ballwin, MO).
Single-Cell Microinjection Assay.
Microinjection assays of coactivator function in Rat-1 fibroblasts were performed essentially as described (8).
Gene Construction.
CFP (mutations relative to wild-type GFP: K26R, F64L, S65T, Y66W, N146I, M153T, V163A, N164H, and H231L) was fused to the N-terminal region of either full-length human RARα or LBDs of human RARα (amino acids 170–462) and human ERα (amino acids 280–595). YFP (mutations S65G, S72A, T203Y, and H231L) was fused to the C-terminal region of the nuclear receptor interaction domain of human SRC-1 (amino acids 623–711) and murine PBP (amino acids 579–651) (see Fig. 2B). All fusions were expressed by using pcDNA3 as vector and contained the N-terminal nuclear localization sequence MPKKKKR.
Pulldown Assays.
We prepared 35S-labeled protein of the GFP chimeras by in vitro transcription and translation as described (24). Glutathione S-transferase (GST)–RAR (residues 20–462) and GST–SRC-1 (residues 623–712) proteins were prepared as crude bacterial lysates, purified over glutathione-agarose beads, and used for a protein–protein interaction assay as described (24).
Transactivation of a RA-Response Element Reporter.
Transient transfection experiments in HeLa cells were performed by using the standard calcium phosphate procedure (25). Cells were transfected with 1 μg of RA-response element-luciferase reporter (DR5) and 1 μg of either Rous sarcoma virus-RAR or pcDNA3-CFP-RAR expression plasmids; the cells were then treated with 0.1 μM TTNPB and assayed for luciferase activity 24 h later.
Live-Cell Imaging and Physiology.
HeLa cells were plated on glass coverslips and transfected with lipofectin (GIBCO/BRL). DNA ratios of the two expression vectors were adjusted to obtain approximately equal amounts of the pair of interacting proteins. Between 12 and 24 h after transfection, cells were imaged at 22°C with a cooled charge-coupled device camera (Photometrics, Tucson, AZ) as described (21). Three successive digital images (with appropriate background subtraction) were acquired at each time point: a FRET image (acceptor YFP emission while exciting the donor CFP: 440 ± 10 nm excitation, 535 ± 12.5 nm emission), a donor CFP image (440 ± 10 nm excitation, 480 ± 15 nm emission), and an acceptor YFP image exciting YFP (495 ± 5 nm excitation, 535 ± 12.5 nm emission). The dichroic mirror used was 455 DRLP for all images. The third image was a reference channel, independent of FRET, to detect possible artifactual effects on YFP such as changes in pH (26), photoisomerization, or defocusing. A binning of two was used to improve the signal/noise ratio. During microscopy, Hanks' balanced salt solution with 25 mM Hepes (pH 7.35) was used as extracellular solution. Agonist stocks were dissolved in 10% DMSO/90% ethanol. The final solvent concentration was 0.1% during addition of compounds.
Photobleaching YFP to Assess Efficiency of FRET.
To quantify the efficiency of FRET in absolute terms, we selectively photobleached the acceptor fluorophore and measured the dequenching of the donor fluorescence (23). FRET efficiency E is given by: E = 1 − (Fda/Fd), where Fda is the donor emission when both donor and acceptor are present, and Fd is donor emission in the absence of acceptor, i.e., after photobleach. We routinely photobleached YFP in cells at the end of each experiment by using a 525 ± 20 nm excitation filter and illuminating the cells for 3–5 min with no neutral density filters. This protocol had no effect on CFP in the absence of YFP.
Results
PBP is Required for RAR and ER Function.
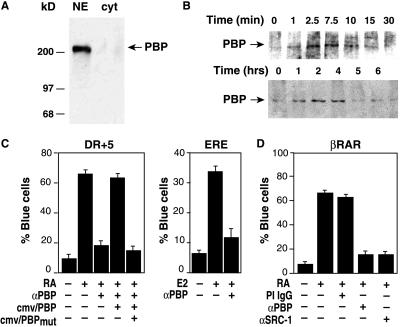
An antibody highly specific for the 220-kDa PBP protein in nuclear extracts (Fig. 1A) was used in low stringency coimmunoprecipitation experiments to assess whether PBP associates with RAR in cells (Fig. 1B). Cells treated with RA for various times were lysed and immunoprecipitated with anti-RAR antibody. The immunoprecipitates were resolved by SDS/PAGE and assayed for associated PBP by Western blotting by using the specific anti-PBP antibody (Fig. 1B). PBP association with RAR depended on RA and was maximal about 2.5 min after RA addition.
Figure 1.
Time course of PBP interactions with RAR and its requirement for RAR- and ER-dependent transcription. (A) Western blot of nuclear and cytoplasmic extracts with anti-PBP antibody, indicating specificity for a 220-kDa band. (B) Time course of PBP interaction with RAR after RA treatment. HeLa cells treated with 1 μM RA were harvested for immunoprecipitation with anti-RAR antibody at the indicated times. Immunoprecipitates were resolved by SDS/PAGE, transferred to nitrocellulose membranes, and probed with anti-PBP antibody. (C) Microinjection of anti-PBP antibody abolishes RA and estrogen-dependent transcription of transiently transfected reporter genes. Rat-1 cells were microinjected with expression vectors for RAR or ER as indicated and the corresponding LacZ reporter genes. Cells were coinjected with control IgG or anti-PBP IgG, treated with agonist, and assayed for LacZ expression 24 h later. Effects of anti-PBP were rescued by overexpression of wild type PBP (cmv/PBP), but not by PBP containing mutations in the LXXLL interaction motifs (cmv/PBPmut). (D) Microinjection of anti-SRC-1 and anti-PBP IgG abolishes RA-dependent transcription in Rat-1 cells containing a chromosomally integrated RAR β2 reporter gene.
To explore whether PBP is a component of an alternative pathway for nuclear receptor activation or is required for ligand-dependent transcription, the effects of microinjected anti-PBP antibodies on an estrogen-dependent transcription were examined (Fig. 1C). Ligand-dependent transcription was initially assayed in Rat-1 fibroblasts microinjected with RAR or ER-responsive reporter genes that directed expression of LacZ. By using these same reporter genes, we have demonstrated that ligand-dependent transcription by RAR and ER requires SRC-1 (14). In cells coinjected with control IgG, treatment with RA increased the percentage of LacZ-expressing cells from 9% to 63%. In contrast, microinjection of anti-PBP reduced the percentage of LacZ-expressing cells to 18%. This inhibition could be rescued by overexpression of wild-type PBP, but not by PBP containing mutations in the LXXLL recognition motifs, suggesting that direct interaction with the nuclear receptor LBD was crucial for function. Microinjection of anti-PBP antibody also blocked ligand-dependent transactivation of a transiently transfected estrogen-dependent reporter gene.
Because the PBP complex does not possess histone acetyltransferase activity and transiently transfected reporter genes may not acquire normal chromatin structure, microinjection experiments were also performed with Rat-1 cells in which LacZ was under transcriptional control of a chromosomally integrated RAR β2 promoter. Microinjection of anti-PBP or anti-SRC-1 antibodies blocked RA-dependent activation of this promoter (Fig. 1C); thus, ligand-dependent transcription of the RAR and ER-dependent promoters requires both SRC-1 and PBP.
Detection of Ligand-Dependent Interaction Between Nuclear Receptors and Coactivators in Cells.
To establish a FRET assay for nuclear receptor-coactivator interaction, we fused CFP to the N terminus of full-length RAR, the RAR LBD or the ER-LBD. YFP was fused to the C-terminal regions of the nuclear receptor interaction domains of SRC-1 or PBP (Fig. 2B). The SRC-1 fragment used for these studies contains LXXLL motifs 1 and 2 and is identical to the SRC-1 peptide cocrystallized with a dimer of the peroxisome proliferator-activated receptor γ (PPARγ) LBD. In this crystal structure, the two LXXLL motifs of a single SRC-1 peptide formed identical contacts with the AF-2 domains of each member of the LBD dimer. This same region of SRC-1 was also found to use both LXXLL motifs to interact cooperatively with DNA-bound RAR/retinoid X receptors heterodimers in vitro (27).
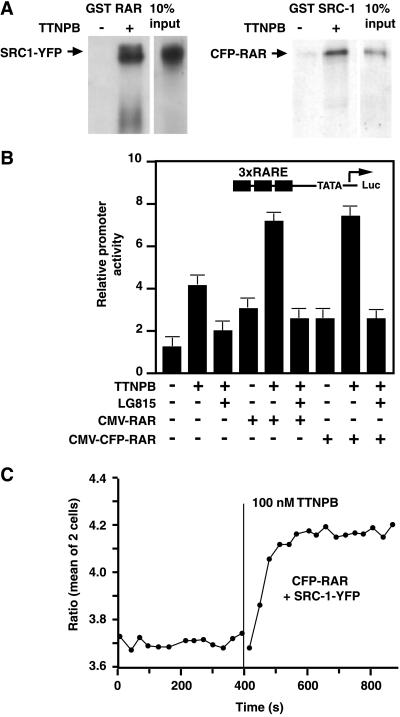
To test whether the tagging of SRC-1 and RAR altered their ability to interact in biochemical assays, we examined the interaction of 35S-labeled CFP–RAR, CFP–RAR LBD, and SRC-1–YFP proteins with appropriate GST-fused partners in vitro. GST–RAR was able to pulldown SRC-1–YFP in a TTNPB-dependent manner (Fig. 3A). Similarly, GST–SRC-1 pulled down CFP–RAR and CFP–RAR LBD ligand dependently (Fig. 3A); thus, the presence of the GFP and YFP moieties in these chimeric proteins did not affect their ability to interact with GST-fused partners in vitro.
Figure 3.
Interactions between CFP–RAR and SRC-1–YFP. (A) In vitro interactions of SRC-1 and RAR fusion proteins monitored by GST pulldown assays. (B) Potentiation of RA-dependent transcription by full-length RAR or the CFP–RAR chimera. HeLa cells were transfected with a RA-dependent reporter gene containing three RA response elements and expression vectors for wild-type RAR or CFP-RAR as indicated. Cells were treated with the RAR agonist TTNPB and the RAR antagonist LG815 as indicated for 24 h before determination of luciferase activity. (C) Imaging the interaction between CFP–RAR full-length and SRC-1-YFP by FRET in live HeLa cells. The ratio of FRET channel emission and donor CFP emission in response to the RAR ligand TTNPB is shown.
To test whether the fusion of CFP to RAR altered its transcriptional properties, HeLa cells were cotransfected with CFP–RAR or wild-type RAR expression vectors, and a RA-dependent reporter gene. The synthetic RAR agonist TTNPB induced transcriptional activity of endogenous RAR, and this activity was potentiated by overexpression of both wild-type RAR and CFP–RAR (Fig. 3B). Therefore, CFP–RAR is a functional receptor, and the presence of CFP at the N terminus of the receptor does not interfere with gene transactivation, similar to results reported for GFP fusions to several nuclear receptors (20).
The subcellular localization of the chimeras was examined by fluorescence microscopy. HeLa cells transiently transfected with CFP–RAR showed nuclear localization of fluorescence with some punctate pattern over a more diffuse nuclear staining (data not shown), as reported for GFP fusions of other full-length nuclear receptors (20). In contrast, the nuclear staining of CFP–RAR LBD and SRC-1–YFP was diffuse (data not shown). Fluorescences of CFP and YFP were therefore spatially averaged in nuclear regions and then ratioed. Fig. 3D shows that when the RAR agonist TTNPB (100 nM) was added to cells expressing both CFP–RAR and SRC-1–YFP, the basal ratio YFP/CFP increased to a new plateau. The increased ratio resulted from dimming of the CFP signal and increased emission of the acceptor YFP (data not shown). This suggests that ligand binding to RAR increases the efficiency of FRET between the GFPs owing to a decrease in the distance or change in orientation between CFP–RAR and SRC-1–YFP. The natural RAR ligand RA produced the same effect as TTNPB, and their effects were not additive (data not shown). However, ligands to receptors other than RAR, such as the ER ligand E2, failed to induce a change in FRET (data not shown).
FRET imaging in live cells is particularly advantageous for monitoring the rate of association, which was rapid. The time constant averaged 33 ± 18 s (mean ± SD for 19 cells) at room temperature, and the association remained constant for at least 2 h in the presence of TTNPB. This rise time may reflect in part the mixing of the ligand in the extracellular solution and is probably an underestimate of the rapidity of RAR/SRC-1 interaction. The ratio change varied from cell to cell and ranged between 5% and 15% (11.3 ± 2.6%, mean ± SD, n = 18 cells). By comparison, the FRET efficiencies measured by photobleaching CFP were 6.1 ± 2.3% before TTNPB, rising to 9.7 ± 2.3% afterward (n = 8). The modest change may reflect either dilution by a large fraction of noninteracting or unlabeled receptors/coactivators, unfavorable orientations of the CFP and YFP chromophores, or a distance between the chromophores significantly greater than 5 nm, the spacing at which FRET is predicted to be 50% efficient between randomly oriented CFP and YFP (28). The biochemical assays for RAR/SRC-1 interaction indicate at most 20% of the CFP–RAR interacted with GST–SRC-1 (Fig. 3A), so it is quite plausible that a majority of CFP–RAR remains uncomplexed to SRC-1–YFP.
Control experiments with single transfection of CFP–RAR or SRC-1–YFP excluded artifactual changes in fluorescence because of alterations in cellular physiology. When cells were cotransfected with unlabelled RAR and SRC-1–YFP, TTNPB also failed to change fluorescence (data not shown). This ruled out the possibility that binding to RAR would change the quantum yield of the YFP, leading to an artifactual change in ratio, independent of FRET.
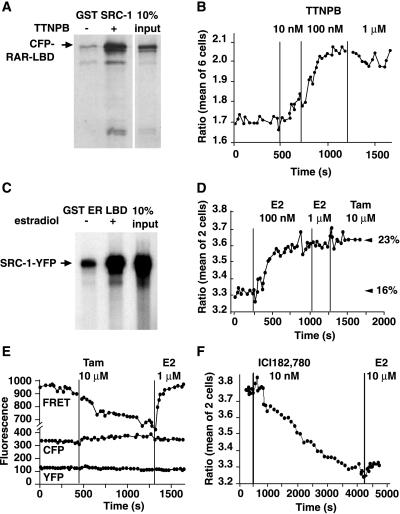
We found virtually identical results when only the LBD of RAR was fused to CFP and the interaction was probed with the same SRC-1–YFP as above (Fig. 4A and B). Addition of TTNPB (as low as 10 nM) increased FRET efficiency, i.e., association between receptor and coactivator; thus, binding to DNA is not necessary for RAR and SRC-1 (1 + 2) to interact in live cells.
Figure 4.
Interaction between SRC-1–YFP and the CFP-tagged LBDs of RAR and ER. In vitro interaction of GST–SRC-1 and CFP–RAR LBD fusion proteins monitored by the GST pulldown assay in the presence or absence of the RAR agonist, TTNPB. (B) Dose-response of the interaction of CFP–RAR LBD and SRC-1–YFP in live cells monitored by FRET. (C) E2 promotes the interaction between GST–ER LBD and SRC-1–YFP in pulldown assays. (D) E2 stimulates FRET in a manner that is resistant to subsequent addition of tamoxifen (Tam). The figures on the right of the panel show the percentage of FRET efficiency calculated by using the YFP-photobleach protocol and indicate ligand-independent interaction in resting cells (16% in the absence of E2 and 23% in its presence). (E) FRET monitoring of CFP–ER LBD and SRC-1–YFP interaction. Tamoxifen inhibits basal FRET in a manner that is reversed by E2. The three channels (FRET, donor CFP, acceptor YFP) are shown. (F) ICI182,780 inhibits basal FRET between CFP–ER LBD and SRC-1–YFP in a manner that cannot be reversed by a 1,000-fold molar excess of E2.
Interactions Between SRC-1 and the ER LBD.
SRC-1 has been shown to be required for transcriptional activation by the ER in Rat-1 fibroblasts (14). Because the LBD of RAR was sufficient for binding SRC-1–YFP in live cells, we constructed the fusion protein CFP–ER LBD to probe the interaction of ER with SRC-1 (Fig. 2B). The ER LBD was considered sufficient because biochemical experiments confirmed that E2 increased the interaction of GST–ER LBD with [35S]SRC-1–YFP (Fig. 4C). The fluorescence in cells transfected with CFP–ER LBD was nuclear as expected (data not shown). In HeLa cells expressing both CFP–ER LBD and SRC-1-YFP, addition of 100 nM E2 increased the ratio YFP/CFP by about 10% (Fig. 4D). Subsequent addition of more E2, or the ER receptor antagonists tamoxifen (10 μM) or ICI182,780 (10 μM) did not have any effect. The FRET efficiency, estimated with the YFP photobleach protocol, increased from 16% basal to 23% after E2.
Because basal FRET was so much higher from CFP–ER LBD to SRC-1–YFP than for CFP–RAR to SRC-1–YFP, SRC-1 and ER LBD seemed to interact in the absence of added estrogen. Fig. 4C confirmed that interaction between GST–ER LBD and [35S]SRC-1–YFP was already noticeable before E2. We then tested whether an ER antagonist could reverse this interaction. Addition of tamoxifen (10 μM) to naive cells expressing the same two chimeric proteins resulted in a decrease in ratio in all cells tested (Fig. 4E; mean 16 ± 4.7%, n = 15 cells), suggesting that there was indeed an interaction between CFP–ER LBD and SRC-1–YFP in the unstimulated cells, which tamoxifen prevented. E2 (1 μM) added after the tamoxifen effect was complete (about 10 min, without washout of tamoxifen) induced a rise in the ratio signal to the basal level before tamoxifen or higher (Fig. 4E). Intriguingly, a 1,000-fold molar excess of E2 was unable to reverse inhibition of FRET by the pure antagonist ICI182,780 (Fig. 4F). This was not caused by degradation of the CFP–ER LBD, because CFP fluorescence was noted to increase after addition of ICI182,780, consistent with the loss of FRET (data not shown). The differences in hysteresis between tamoxifen and ICI182,780 suggest that they induce distinct conformational changes in the ER LBD that alter associations with other cellular proteins, such as corepressors.
The changes in ratio by tamoxifen and E2 in these experiments are likely changes in FRET efficiency because the FRET channel signal (sensitized emission of the acceptor YFP) and the donor channel signal (CFP) changed reciprocally (Fig. 4E), whereas direct excitation of the YFP showed no change. The FRET efficiency was calculated to be 16% basal, decreased to 7% by tamoxifen, and was reversed by E2 (Fig. 4E). The basal interaction of ER LBD and SRC-1 (1 + 2) was not caused by estrogen present in the tissue culture medium, because use of charcoal-stripped serum and phenol red-free media did not affect the decrease in ratio by tamoxifen. Finally, when tamoxifen was added to cells after the addition of E2, it was unable to inhibit FRET (Fig. 4B), even when estrogen was washed out and tamoxifen was added at a high molar excess (data not shown).
Interactions Among PBP, RAR, and ER LBD Monitored by FRET.
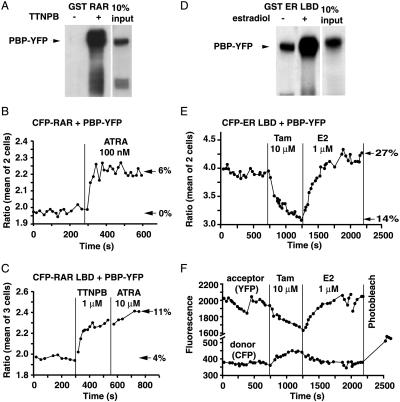
Biochemical studies indicate that PBP and SRC-1 compete with each other for interaction with the same coactivator binding site on the LBD (8, 29). These studies and the microinjection experiments illustrated in Fig. 1 indicate that the coactivator function of both SRC-1 and PBP requires the integrity of the LXXLL interaction motifs, suggesting that these motifs mediate direct interactions with nuclear receptors in cells. To examine whether PBP also interacts directly with the RAR within cells, the region of PBP containing the two consensus LXXLL sequences was fused to YFP at the C terminus (Fig. 2, PBP–YFP). This fusion protein was capable of interacting with GST–RAR and GST–ER in a ligand-dependent manner as shown in Fig. 5 A and D. As with SRC-1, PBP in the absence of ligands clearly interacted with ER LBD but not RAR. In HeLa cells cotransfected with CFP–RAR and PBP–YFP, TTNPB or RA increased the ratio YFP/CFP emission by about 10.7 ± 2.5% (n = 8 cells), whereas FRET efficiency increased from 0 to 6% (Fig. 5B). This interaction was also observed when the LBD of RAR was used (Fig. 5C). The kinetics of the interaction of PBP–YFP and SRC-1–YFP with CFP–RAR in cells were similar.
Figure 5.
PBP interactions with RAR and ER. (A) GST pulldown assay demonstrating ligand-dependent interaction between PBP–YFP and GST–RAR or GST–ER LBD. (B and C) Emission ratio detection of FRET from CFP–RAR full-length (B) or CFP–RAR LBD (C) to PBP–YFP on addition of RA or the synthetic RAR ligand TTNPB. The figures on the right of the panels show the percentage of FRET efficiency calculated by using the YFP-photobleach protocol at the end of each experiment. (D) GST pulldown assay demonstrating ligand-dependent interaction between PBP–YFP and GST–ER LBD. (E) Emission ratio monitoring of the interaction between CFP–ER LBD and PBP–YFP. Tamoxifen reduces the interaction, and estrogen promotes it. (F) The fluorescence intensity of FRET and donor channels for the above experiment is shown. The artifact at around 400 s represents refocusing, which cancels out when the YFP and CFP are ratioed. The CFP fluorescence after photobleaching YFP is Fd.
We next probed the interaction of PBP with ER in cells. As with ER/SRC-1, the ER LBD exhibited a basal interaction with PBP (Fig. 5E). Cells expressing CFP–ER LBD and PBP–YFP showed a drop in the ratio YFP/CFP on addition of tamoxifen (10 μM). E2 (0.1 or 1 μM) added in the presence of tamoxifen raised the ratio to the resting level or above (Fig. 5E), depending on the cell. Photobleaching confirmed the existence of basal ligand-independent interaction (Fig. 5F).
Discussion
We have monitored the association of SRC-1 and PBP-interaction domains with ER and RAR in living cells by using FRET and demonstrated by antibody microinjection that both SRC-1 and PBP are required for ligand-dependent transactivation from transiently transfected and chromosomally integrated promoters. Thus, rather than being redundant factors or components of distinct transcriptional activation pathways, SRC-1 and PBP serve essential and complementary transcriptional roles, at least for the promoters used in these studies, consistent with the distinct biochemical properties of SRC-1 and PBP and their associated complexes.
The ability of PBP to function as a coactivator in microinjection experiments required that it possess intact LXXLL recognition motifs that mediate interactions with nuclear receptor LBDs in vitro. Furthermore, FRET assays demonstrated that both the SRC-1 and PBP nuclear receptor interaction domains could interact with the RAR and ER in cells with similar rapid kinetics. Because PBP and SRC-1 each use LXXLL motifs to interact with the nuclear receptor AF-2 domains, they cannot simultaneously interact with the same receptor. At least two models could explain the apparent paradox that both PBP and SRC-1 are required for activation by ER and RAR, yet only one of these proteins can be bound to a receptor at a time. First, the promoters used in these studies contain multiple binding sites for ER or RARs. Assuming that more than one receptor dimer or heterodimer binds to these elements in cells, the possibility exists that SRC and PBP complexes are simultaneously recruited to adjacent receptors. The coordinate recruitment of two complexes with distinct activities could potentially account for the self-synergy that is obtained when multiple response elements are placed upstream of a minimal promoter. Alternatively, the observations reported here are also consistent with a model in which there is a sequence of coactivator recruitment involving the exchange of SRC-1 and PBP-containing complexes. In this model, it is most plausible to suggest that recruitment of SRC-1 complexes would occur first, resulting in histone acetylation and chromatin remodeling that renders the promoter more accessible to core factors. The SRC-1 complex would then be replaced by PBP-containing complexes that would direct assembly of core factors and promote transcriptional initiation. An ordered sequence of recruitment of transcription factors and coactivators to a regulated promoter has recently been documented during the cell cycle-dependent activation of the HO gene in yeast (30).
At present, our studies do not discriminate between these two models, and other explanations are, of course, possible. The ability to monitor ligand-dependent interactions in cells on a time scale of seconds suggests that it may be possible to obtain evidence for sequential interactions of nuclear receptors and coactivators. Although the kinetics of ligand-dependent interaction of SRC-1 and PBP with RAR and ER were very similar, these experiments used the nuclear receptor interaction domains of SRC-1 and PBP, rather than full-length proteins. The interactions of these domains with liganded ER and RAR were very stable, and in the case of ER LBD experiments, could not be disrupted with tamoxifen. Thus, if an exchange of SRC-1-containing complexes occurs with PBP or other factors, an active mechanism must be needed to disengage SRC-1. Acetylation of conserved Lys residues in the vicinity of the LXXLL interaction motifs of p/CIP/ACTR and SRC-1 has been suggested to inhibit nuclear receptor interaction and could potentially represent a signal that would initiate the exchange of a p160 factor for another coactivator protein (31). Such modification of p160 proteins would presumably require the recruitment of associated factors such as CBP and pCAF that harbor histone acetyltransferase activities, which would not be expected to occur with the SRC-1–YFP fusion proteins used for FRET experiments in these studies. However, the ability to measure ligand-dependent interactions between full-length RAR and the nuclear receptor interaction domains of SRC-1 and PBP suggest that experiments with larger coactivator fragments and eventually with full-length proteins will be feasible. These approaches are likely to lead to new insights into mechanisms of coactivator assembly and function in living cells.
Acknowledgments
This work was supported by National Institutes of Health Grants NS27177 to R.Y.T. and CA52599 to C.K.G., and Office of Naval Research, Order No. N00014–98-F-0402 to the Molecular Design Institute through the U.S. Department of Energy under Contract No. DE-AC03–76SF00098 (J.L. and R.Y.T.). M.R. is supported by a postdoctoral fellowship from the Western States Affiliates of the American Heart Association.
Abbreviations
- TTNPB
(4-[(E)-2-(5, 6, 7, 8-tetrahydro-5, 5, 8, 8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid
- ER
estrogen receptor
- RA
retinoic acid
- RAR
retinoic acid receptor
- LBD
ligand-binding domain
- FRET
fluorescence resonance energy transfer
- E2
17β-estradiol
- GFP
green fluorescent protein
- CFP
cyan fluorescent protein
- YFP
yellow fluorescent protein
- GST
glutathione S-transferase
- SRC
steroid receptor coactivator
- PBP
peroxisome proliferator-activated receptor binding protein
Footnotes
References
- 1.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glass C K. Endocr Rev. 1994;15:1503–1519. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 3.Moras D, Gronemeyer H. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 4.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engström O, Öhman L, Greene G L, Gustafsson J-Å, Carlquist M. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 5.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 6.Voegel J J, Heine M J, Tini M, Vivat V, Chambon P, Gronemeyer H. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 8.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 9.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 10.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 11.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oñate S A, Tsai S Y, Tsai M-J, O'Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 13.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R, Rose D, Glass C, et al. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 14.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 15.McKenna N J, Lanz R B, O'Malley B W. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 16.Rachez C, Suldan Z, Ward J, Chang C-P B, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fondell J D, Ge H, Roeder R G. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rachez C, Lemon B, Suldan Z, Bromleigh V, Gamble M, Näär A, Erdjument-Bromage H, Tempst P, Freedman L. Nature (London) 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 19.Yuan C-X, Ito M, Fondell J D, Fu Z-Y, Roeder R G. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Htun H, Holth L T, Walker D, Davie J R, Hager G L. Mol Biol Cell. 1999;10:471–486. doi: 10.1091/mbc.10.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyawaki A, Llopis J, McCaffery J M, Adams J A, Ikura M, Tsien R Y. Nature (London) 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 22.Day R N. Mol Endocrinol. 1998;12:1410–1419. doi: 10.1210/mend.12.9.0168. [DOI] [PubMed] [Google Scholar]
- 23.Miyawaki, A. & Tsien, R. Y. (2000) Methods Enzymol., in press. [DOI] [PubMed]
- 24.Kurokawa R, Kalafus D, Ogliastro M-H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 25.Ricote M, Li A C, Willson T M, Kelly C J, Glass C K. Nature (London) 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 26.Llopis J, McCaffery J M, Miyawaki A, Farquhar M G, Tsien R Y. Proc Natl Acad Sci USA. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Nature (London) 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 28.Heim R. Methods Enzymol. 1999;302:408–423. doi: 10.1016/s0076-6879(99)02036-4. [DOI] [PubMed] [Google Scholar]
- 29.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T-M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, et al. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cosma M P, Tanaka T, Nasmyth K. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 31.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]