Abstract
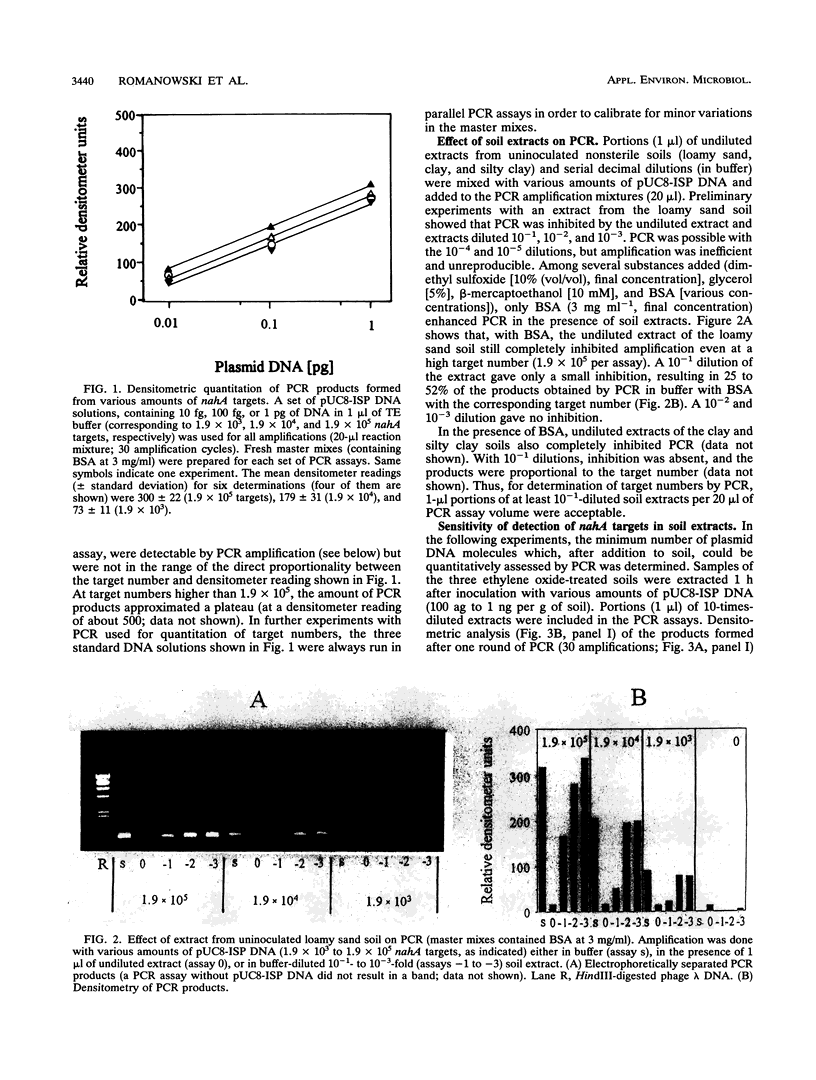
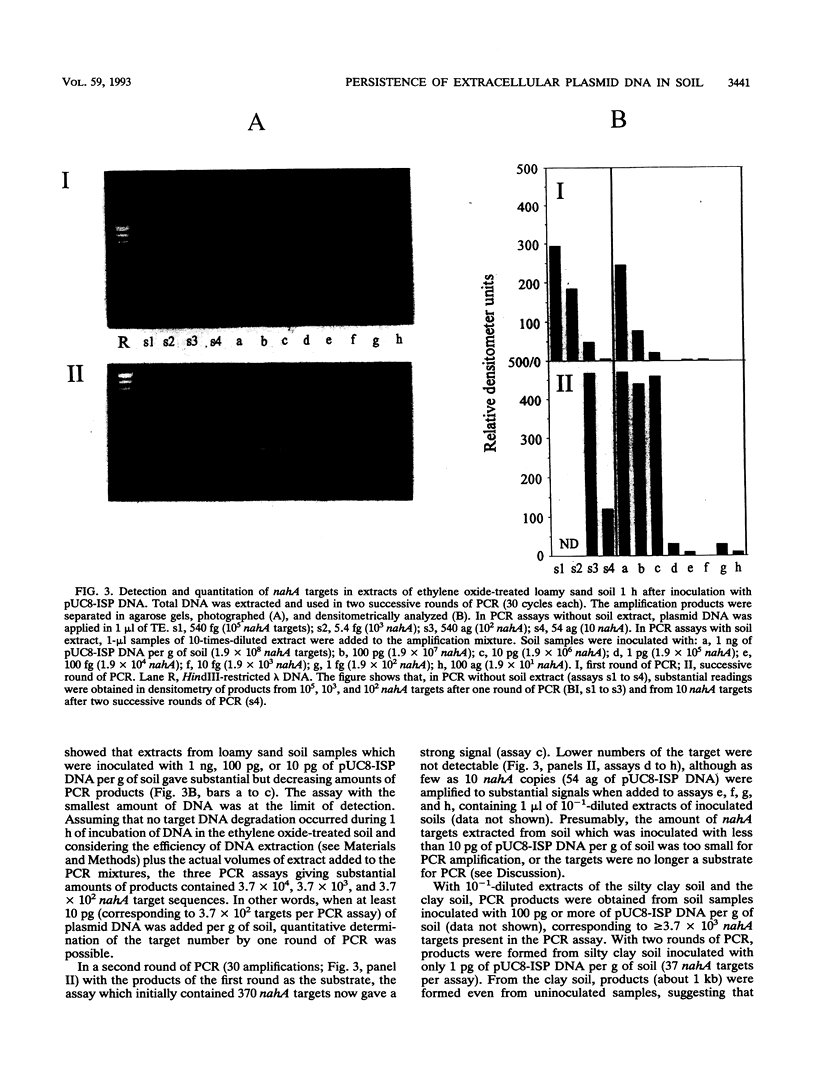
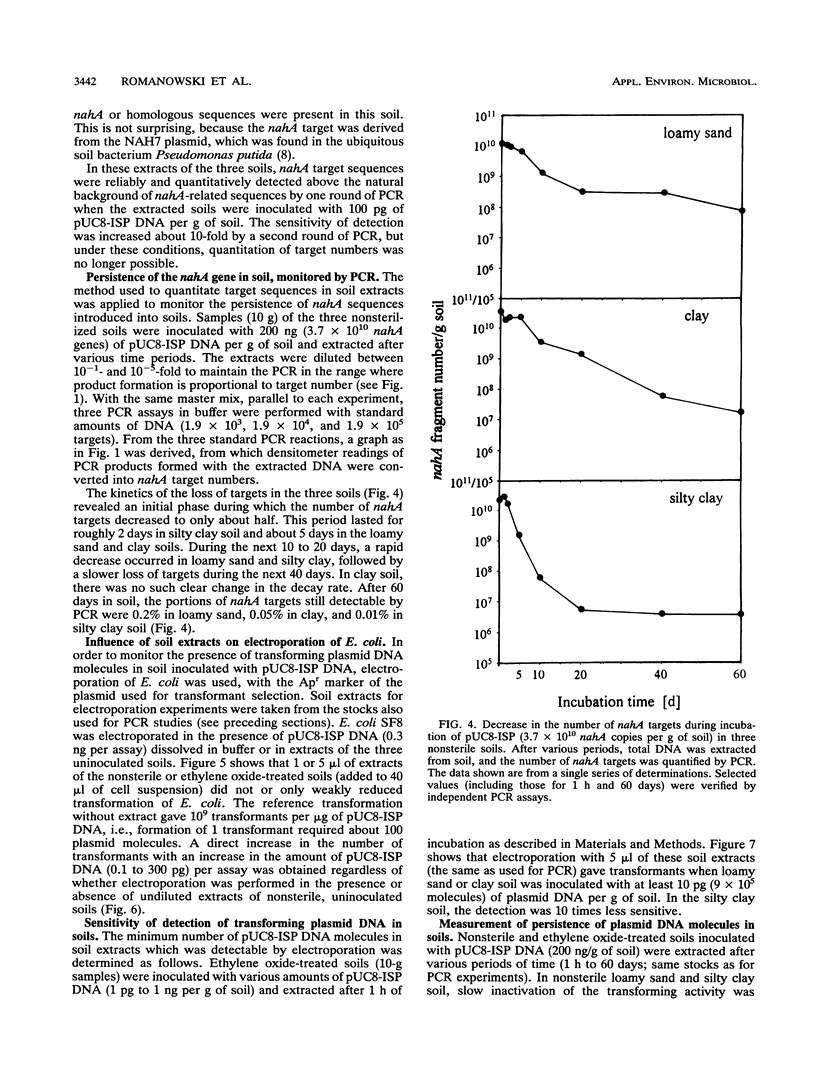
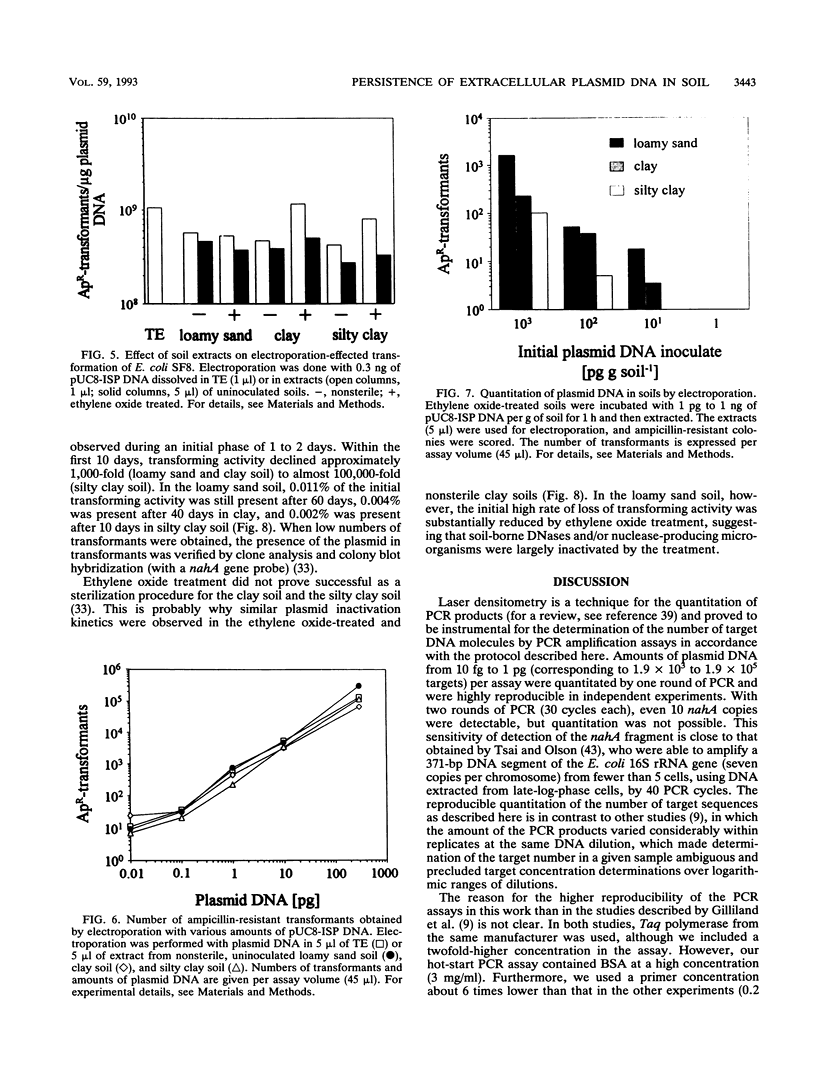
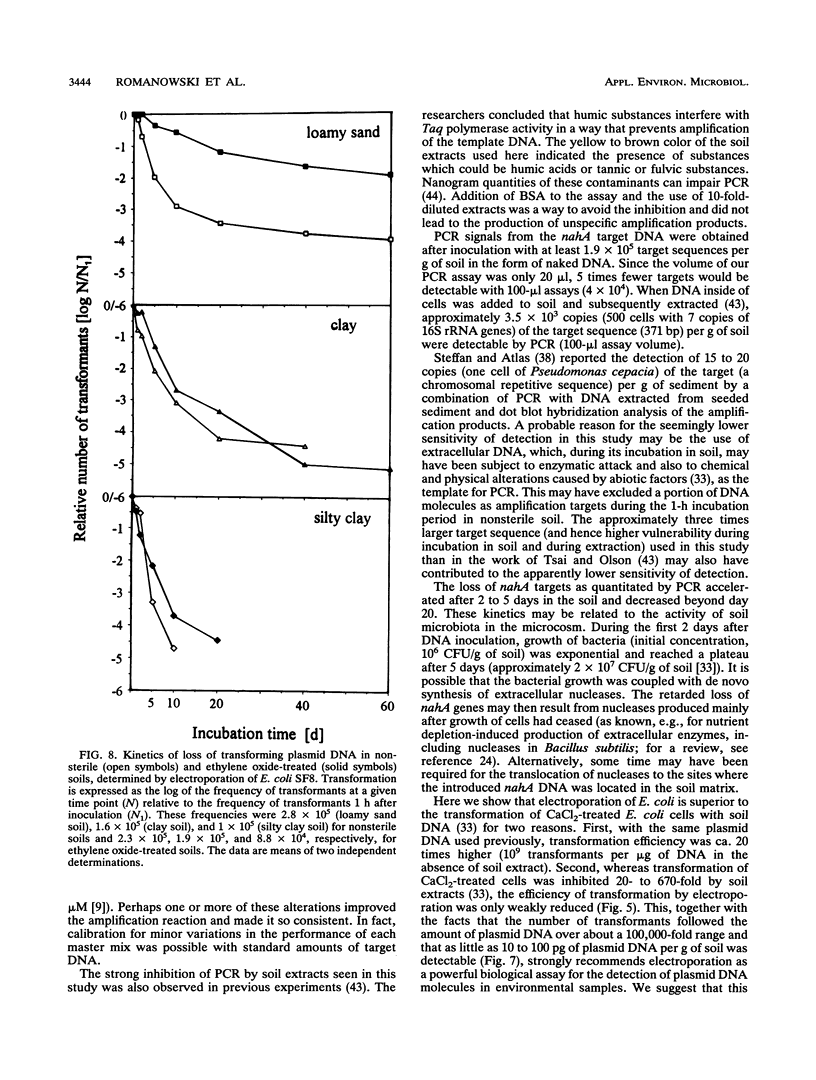
A modified protocol for DNA amplification by polymerase chain reaction (PCR) coupled with laser densitometric determination of the amount of PCR products, which allowed quantitation of target sequence numbers in soil extracts, was developed. The method was applied to monitor target loss during incubation of purified plasmid DNA in natural nonsterile soils. It revealed soil-specific kinetics of target loss. After 60 days, 0.2, 0.05, and 0.01% of the initially added nahA genes on plasmids were detectable by PCR in a loamy sand soil, a clay soil, and a silty clay soil, respectively. Electroporation of Escherichia coli was used in parallel to quantitate plasmid molecules in soil extracts by their transforming activity. It was found that transformation by electroporation was about 20 times more efficient and much less inhibited by constituents of soil extracts than transformation of Ca(2+)-treated cells (G. Romanowski, M.G. Lorenz, G. Sayler, and W. Wackernagel, Appl. Environ. Microbiol. 58:3012-3019, 1992). By electroporation, greater than 10,000-fold plasmid loss was monitored in nonsterile soils. Transforming activity was found up to 60 days after inoculation of the soils. The studies indicate that PCR and electroporation are sensitive methods for monitoring the persistence of extracellular plasmid DNA in soil. It is proposed that plasmid transformation by electroporation can be used for the monitoring in soil and other environments of genetically engineered organisms with recombinant plasmids. The data suggest that genetic material may persist in soil for weeks and even for months after its release from cells.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bien M., Steffen H., Schulte-Frohlinde D. Repair of the plasmid pBR322 damaged by gamma-irradiation or by restriction endonucleases using different recombination-proficient E. coli strains. Mutat Res. 1988 Nov;194(3):193–205. doi: 10.1016/0167-8817(88)90021-1. [DOI] [PubMed] [Google Scholar]
- CATLIN B. W. Extracellular deoxyribonucleic acid of bacteria and a deoxyribonuclease inhibitor. Science. 1956 Sep 7;124(3219):441–442. doi: 10.1126/science.124.3219.441. [DOI] [PubMed] [Google Scholar]
- Chamier B., Lorenz M. G., Wackernagel W. Natural Transformation of Acinetobacter calcoaceticus by Plasmid DNA Adsorbed on Sand and Groundwater Aquifer Material. Appl Environ Microbiol. 1993 May;59(5):1662–1667. doi: 10.1128/aem.59.5.1662-1667.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb W. D., Streips U. N., Doyle R. J. Selective enrichment for genetic markers in DNA released by competent cultures of Bacillus subtilis. Mol Gen Genet. 1977 Oct 20;155(2):179–183. doi: 10.1007/BF00393157. [DOI] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland G., Perrin S., Blanchard K., Bunn H. F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Khanna M., Stotzky G. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl Environ Microbiol. 1992 Jun;58(6):1930–1939. doi: 10.1128/aem.58.6.1930-1939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. G., Aardema B. W., Wackernagel W. Highly efficient genetic transformation of Bacillus subtilis attached to sand grains. J Gen Microbiol. 1988 Jan;134(1):107–112. doi: 10.1099/00221287-134-1-107. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Gerjets D., Wackernagel W. Release of transforming plasmid and chromosomal DNA from two cultured soil bacteria. Arch Microbiol. 1991;156(4):319–326. doi: 10.1007/BF00263005. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Reipschläger K., Wackernagel W. Plasmid transformation of naturally competent Acinetobacter calcoaceticus in non-sterile soil extract and groundwater. Arch Microbiol. 1992;157(4):355–360. doi: 10.1007/BF00248681. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Appl Environ Microbiol. 1987 Dec;53(12):2948–2952. doi: 10.1128/aem.53.12.2948-2952.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. High Frequency of Natural Genetic Transformation of Pseudomonas stutzeri in Soil Extract Supplemented with a Carbon/Energy and Phosphorus Source. Appl Environ Microbiol. 1991 Apr;57(4):1246–1251. doi: 10.1128/aem.57.4.1246-1251.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Natural genetic transformation of Pseudomonas stutzeri by sand-adsorbed DNA. Arch Microbiol. 1990;154(4):380–385. doi: 10.1007/BF00276535. [DOI] [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Paul J. H., David A. W. Production of extracellular nucleic acids by genetically altered bacteria in aquatic-environment microcosms. Appl Environ Microbiol. 1989 Aug;55(8):1865–1869. doi: 10.1128/aem.55.8.1865-1869.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Frischer M. E., Thurmond J. M. Gene transfer in marine water column and sediment microcosms by natural plasmid transformation. Appl Environ Microbiol. 1991 May;57(5):1509–1515. doi: 10.1128/aem.57.5.1509-1515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Jeffrey W. H., DeFlaun M. F. Dynamics of extracellular DNA in the marine environment. Appl Environ Microbiol. 1987 Jan;53(1):170–179. doi: 10.1128/aem.53.1.170-179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Thurmond J. M., Frischer M. E., Cannon J. P. Intergeneric natural plasmid transformation between E. coli and a marine Vibrio species. Mol Ecol. 1992 May;1(1):37–46. doi: 10.1111/j.1365-294x.1992.tb00153.x. [DOI] [PubMed] [Google Scholar]
- Paul John H., Deflaun Mary F., Jeffrey Wade H., David Andrew W. Seasonal and Diel Variability in Dissolved DNA and in Microbial Biomass and Activity in a Subtropical Estuary. Appl Environ Microbiol. 1988 Mar;54(3):718–727. doi: 10.1128/aem.54.3.718-727.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Sayler G., Wackernagel W. Persistence of free plasmid DNA in soil monitored by various methods, including a transformation assay. Appl Environ Microbiol. 1992 Sep;58(9):3012–3019. doi: 10.1128/aem.58.9.3012-3019.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Wackernagel W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl Environ Microbiol. 1991 Apr;57(4):1057–1061. doi: 10.1128/aem.57.4.1057-1061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Wackernagel W. Plasmid DNA in a groundwater aquifer microcosm--adsorption, DNAase resistance and natural genetic transformation of Bacillus subtilis. Mol Ecol. 1993 Jun;2(3):171–181. doi: 10.1111/j.1365-294x.1993.tb00106.x. [DOI] [PubMed] [Google Scholar]
- Sayler G. S., Lund L. C., Shiaris M. P., Sherrill T. W., Perkins R. E. Comparative effects of Aroclor 1254 (polychlorinated biphenyls) and phenanthrene on glucose uptake by freshwater microbial populations. Appl Environ Microbiol. 1979 May;37(5):878–885. doi: 10.1128/aem.37.5.878-885.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- Stewart G. J., Sinigalliano C. D. Detection of horizontal gene transfer by natural transformation in native and introduced species of bacteria in marine and synthetic sediments. Appl Environ Microbiol. 1990 Jun;56(6):1818–1824. doi: 10.1128/aem.56.6.1818-1824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992 Feb;58(2):754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol. 1992 Jul;58(7):2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk V., Rehnstam A. S., Lundberg E., Hagström A. Release of bacterial DNA by marine nanoflagellates, an intermediate step in phosphorus regeneration. Appl Environ Microbiol. 1992 Nov;58(11):3744–3750. doi: 10.1128/aem.58.11.3744-3750.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]