Abstract
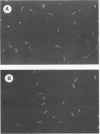
Vibrio vulnificus was enumerated on membrane filters after hybridization with a fluorescent oligonucleotide eubacterial probe. Cells were hybridized in liquid buffer or directly on membrane filters. There was no significant difference between fluorescent oligonucleotide direct counts and acridine orange direct counts (P > 0.05). Liquid buffer hybridization was preferable to direct filter hybridization.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990 Jun;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Stromley J., Devereux R., Key R., Stahl D. A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992 Feb;58(2):614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Chen D., Hanna P. J., Altmann K., Smith A., Moon P., Hammond L. S. Development of monoclonal antibodies that identify Vibrio species commonly isolated from infections of humans, fish, and shellfish. Appl Environ Microbiol. 1992 Nov;58(11):3694–3700. doi: 10.1128/aem.58.11.3694-3700.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks R. E., Amann R. I., Stahl D. A. Dual staining of natural bacterioplankton with 4',6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992 Jul;58(7):2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie J. E., Daley R. J., Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 May;33(5):1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben W. E., Schroeter B. M., Calabrese V. G., Olsen R. H., Kukor J. K., Biederbeck V. O., Smith A. E., Tiedje J. M. Gene probe analysis of soil microbial populations selected by amendment with 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol. 1992 Dec;58(12):3941–3948. doi: 10.1128/aem.58.12.3941-3948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A., Liesack W., Stackebrandt E. Polymerase chain reaction-gene probe detection system specific for pathogenic strains of Yersinia enterocolitica. J Clin Microbiol. 1992 Aug;30(8):1942–1947. doi: 10.1128/jcm.30.8.1942-1947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight I. T., Shults S., Kaspar C. W., Colwell R. R. Direct detection of Salmonella spp. in estuaries by using a DNA probe. Appl Environ Microbiol. 1990 Apr;56(4):1059–1066. doi: 10.1128/aem.56.4.1059-1066.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehnstam A. S., Norqvist A., Wolf-Watz H., Hagström A. Identification of Vibrio anguillarum in fish by using partial 16S rRNA sequences and a specific 16S rRNA oligonucleotide probe. Appl Environ Microbiol. 1989 Aug;55(8):1907–1910. doi: 10.1128/aem.55.8.1907-1910.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. B., Tiedje J. M. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl Environ Microbiol. 1992 Jan;58(1):376–384. doi: 10.1128/aem.58.1.376-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor P. S., Neihof R. A. Improved method for determination of respiring individual microorganisms in natural waters. Appl Environ Microbiol. 1982 Jun;43(6):1249–1255. doi: 10.1128/aem.43.6.1249-1255.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Hanson R. S. Soluble methane monooxygenase component B gene probe for identification of methanotrophs that rapidly degrade trichloroethylene. Appl Environ Microbiol. 1992 Mar;58(3):953–960. doi: 10.1128/aem.58.3.953-960.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]