Abstract
The ability of organisms to acquire thermotolerance to normally lethal high temperatures is an ancient and conserved adaptive response. However, knowledge of cellular factors essential to this response is limited. Acquisition of thermotolerance is likely to be of particular importance to plants that experience daily temperature fluctuations and are unable to escape to more favorable environments. We developed a screen, based on hypocotyl elongation, for mutants of Arabidopsis thaliana that are unable to acquire thermotolerance to high-temperature stress and have defined four separate genetic loci, hot1–4, required for this process. hot1 was found to have a mutation in the heat shock protein 101 (Hsp101) gene, converting a conserved Glu residue in the second ATP-binding domain to a Lys residue, a mutation that is predicted to compromise Hsp101 ATPase activity. In addition to exhibiting a thermotolerance defect as assayed by hypocotyl elongation, 10-day-old hot1 seedlings were also unable to acquire thermotolerance, and hot1 seeds had greatly reduced basal thermotolerance. Complementation of hot1 plants by transformation with wild-type Hsp101 genomic DNA restored hot1 plants to the wild-type phenotype. The hot mutants are the first mutants defective in thermotolerance that have been isolated in a higher eukaryote, and hot1 represents the first mutation in an Hsp in any higher plant. The phenotype of hot1 also provides direct evidence that Hsp101, which is required for thermotolerance in bacteria and yeast, is also essential for thermotolerance in a complex eukaryote.
The ability to acquire tolerance to normally lethal high temperatures is a property of virtually all organisms (1). This type of adaptation can increase the temperature of lethality by one to several degrees centigrade. Even prokaryotic and eukaryotic thermophiles, with optimal growth temperatures of over 60°C, exhibit the ability to acquire thermotolerance (2). The acquisition of thermotolerance results from prior exposure to a conditioning pretreatment, which can be a sublethal high temperature or a number of other moderate stress treatments (1). The ancient origin and evolutionary conservation of this adaptive response suggest that it is a fundamentally important aspect of the interaction of an organism with the abiotic environment.
Despite the ubiquitous nature of the acquired thermotolerance phenomenon, only a limited number of factors have been defined that contribute to the development of thermotolerance. The best-characterized essential factor, in both prokaryotes and eukaryotes, is a protein of the heat shock protein 100 (Hsp100)/ClpB family; these proteins are a class of molecular chaperones found in bacteria and the cytosol of Saccharomyces cerevisiae, higher plants, and some protozoa, but not in other eukaryotes (3). They are part of a large family of ATPases, with conserved P-loop and DExx motifs required for ATPase activity (4). In S. cerevisiae, deletion of the gene encoding the Hsp100/ClpB protein, Hsp104, reduces the ability of cells to survive high-temperature exposure by three to four orders of magnitude (5). Similarly, antisense inhibition of Hsp101 expression in Arabidopsis thaliana severely compromises thermotolerance of seedlings directly after germination or 14 days of growth (37). Hsp100/ClpB is also essential for thermotolerance in a cyanobacterium (6) and plays a role in stress tolerance in other bacteria (3). In vivo and in vitro data indicate that Hsp100/ClpB facilitates the dissolution of protein aggregates in cooperation with other chaperones, including Hsp70 (or DnaK) and Hsp40 (or DnaJ) (7–10). This unique function presumably explains its importance to survival and recovery from heat stress.
Although some other factors required for thermotolerance in addition to Hsp100/ClpB proteins have been defined, their importance may be more organism-specific. In S. cerevisiae, mutants of bcy1, the regulatory subunit of cAMP-dependent protein kinase, exhibit altered cAMP levels, do not arrest in the G1 phase of the cell cycle in response to heat shock and are unable to adapt to high temperature (11). Accumulation of trehalose during stress, as well as activity of the plasma membrane ATPase are also important for thermotolerance in yeast (12). Synthesis of a small cytoplasmic RNA (300 nt), which has restricted homology to 7SL and 4.5 S RNA and appears to associate with ribosomes, has been genetically linked to development of temperature tolerance in Tetrahymena thermophila (13). Small Hsps are essential for thermotolerance in Neurospora crassa (14) and a cyanobacterium (ref. 15 and E.V., unpublished results), but they appear to be dispensable for this function in yeast and other bacteria (16, 17). Although other Hsps may also contribute to thermotolerance, in general they appear to be important for growth at higher temperatures rather than necessary for the specific adaptation processes involved in acquired thermotolerance (18).
Our minimal understanding of the mechanism by which organisms acquire thermotolerance arises in part from a lack of systematic genetic studies dissecting the components involved. In higher eukaryotes, work of this type has been inhibited by the fact that screening procedures to identify loss-of-function mutants result in death of the desired mutants. In model microorganisms, such as S. cerevisiae and Escherichia coli, thermotolerance changes dramatically with growth phase; stationary cells are highly thermotolerant compared with cells in early exponential phase (19). This has prohibited standard genetic screens dependent on plating assays, in which the cell growth phase is variable and not controlled.
Because plants are sessile organisms, with a limited ability to thermoregulate that is highly dependent on water availability (20), rapid adaptation to daily temperature fluctuations is likely essential for their survival. Plants can be adapted to normally lethal temperatures either by short pretreatments at elevated nonlethal temperatures, or by a gradual increase to temperatures that would be lethal if imposed abruptly (21). Thus, during hotter seasons, plants may undergo a daily cycle of acquiring thermotolerance to maintain optimal growth. To test the importance of acquired thermotolerance in plant growth, as well as to define the potentially multiple factors involved in thermotolerance in other eukaryotes, we devised a quantitative assay for thermotolerance of Arabidopsis seedlings that allowed recovery of loss-of-function thermotolerance mutants. With this assay, we have now defined four genetic loci that are necessary for acquired thermotolerance. One locus, hot1 (sensitive to hot temperatures), was found to encode the Arabidopsis Hsp100/ClpB protein Hsp101, representing, we believe, the first mutation in an Hsp gene in any higher plant.
Materials and Methods
Mutant Screen.
For the hypocotyl elongation mutant screen, ethyl methanesulfonate-mutagenized M2 seeds (Col-O ecotype) were purchased from Lehle Seeds (Round Rock, TX). Approximately 17,000 M2 seedlings were screened, representing an estimated 2,100 M1 parents. Surface-sterilized seeds were plated in rows on 10 ml of minimal medium (22) in 10-cm square plates, which were wrapped in foil and incubated at 4°C for a minimum of 3 days. Plates were then placed in a vertical position at 22°C for 2.5 days, at which time they were treated at 38°C for 90 min followed by 2 h at 22°C and then 2 h at 45°C, and then briefly opened to mark the position of the cotyledons. After an additional 2.5 days in the dark, seedlings that showed no further growth were marked for later rescue. Rescued seedlings were transplanted to soil and M3 seed was recovered. M3 seed was retested for hypocotyl elongation under conditions shown in Fig. 1. Only those mutants that failed to grow after a 38°C pretreatment followed by a 45°C heat stress, but that showed normal growth after a 38°C pretreatment, were further analyzed.
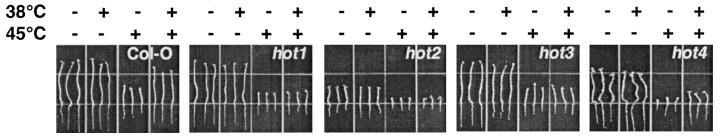
Figure 1.
Hypocotyl elongation phenotype of Arabidopsis seedlings of wild type (Col-O) and four mutants (hot1–4). After growth for 2.5 days in the dark at 22°C, seedlings were either maintained at 22°C; treated at 38°C for 90 min; at 45°C for 2 h; or at 38°C for 90 min followed by 2 h at 22°C and then 45°C for 2 h. Seedlings were returned to 22°C for 2.5 days and then photographed. Groups of three seedlings that received the same treatment, as indicated above the panel, are shown. hot1, -2, and -4 seedlings are F3 progeny of a single backcross to the wild-type parent. hot3 seedlings are from the M3 generation, but show an identical phenotype to mutants segregating in the F2 progeny of a backcross (not shown).
Genetic Analyses.
M3 plants of defined mutants were backcrossed to the wild-type Col-O parent and outcrossed to the Landsberg erecta ecotype for mapping by using the mutant as the female parent. All plants used for crosses were maintained in a growth chamber under continuous light at 24°C. F1 seedlings from the backcross were analyzed in the hypocotyl elongation assay to determine dominance. F2 seedlings from the backcross were used to assess segregation of the phenotype in the hypocotyl assay. Mutants were analyzed for complementation in reciprocal crosses by testing of the F1 progeny in the hypocotyl assay.
For mapping of the hot1 mutation, 48 F2 seedlings showing the mutant phenotype were isolated from the outcrossed F2 population. Genomic DNA was extracted according to Klimyuk et al. (23). The DNA was used in PCRs to score the cosegregation of the mutant phenotype with simple sequence-length polymorphic markers (24).
Vector Construction and Plant Transformation.
A 6,609-kb XbaI fragment of Landsberg erecta genomic DNA, spanning the AtHsp101 gene (E.V., unpublished results), was cloned into the XbaI site of the pBin19 vector (25). The Hsp101 gene or the vector alone were transformed into hot1 mutant plants by a modified vacuum infiltration method (26). Transformed seedlings were selected on minimal plates (22) with kanamycin (30 μg/ml) for 10 days before transfer to soil. Progeny from the kanamycin-resistant plants were tested for acquired thermotolerance in the hypocotyl assay.
DNA Sequence Analysis.
The region of genomic DNA used for complementation of the hot1 mutant (Fig. 2) was amplified by PCR from Col-O wild-type or hot1 mutant plants by using primers designed to give overlapping fragments for DNA sequencing, The PCR fragments were separated by electrophoresis, excised, and the DNA was purified with a Qiaex II gel kit (Qiagen, Chatsworth, CA). Sequencing was performed on an ABI 377 sequencer by the University of Arizona Biotechnology Facility by using the PCR primers. Sequence analysis was conducted with the genejockey ii program (Biosoft, Cambridge, U.K.).
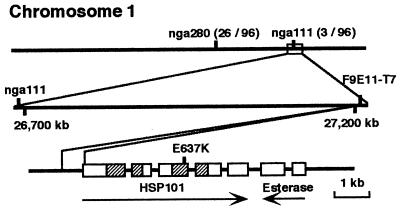
Figure 2.
hot1 has a missense mutation in a conserved residue in the second ATP-binding domain of Arabidopsis Hsp101. The upper line shows a segment of the lower arm of chromosome 1 with simple sequence-length polymorphism markers nga 280 and nga111. The number of recombinants recovered between these markers and the hot1 mutation, per chromosome screened, are indicated in parentheses. The second line shows an expansion of the region adjacent to nga111 with the position of bacterial artificial chromosome F9E11, whose T7 end sequence is identical to ≈570 bp the Hsp101 gene, including ≈200 bp of coding region. The third line shows the structure of the Hsp101 gene and neighboring putative esterase gene. Boxes indicate exons and hatched boxes indicate the ATP-binding domains of Hsp101. Position of the E → K mutation in hot1 is indicated.
Other Thermotolerance Assays.
For all assays, seeds were first surface sterilized and planted on minimal plates (22), which were then wrapped in foil and incubated at 4°C for 3 days. To test seed basal thermotolerance, seeds were heat stressed for 2 h at 45°C immediately on removal from the cold. They were then measured and photographed after an additional 3 days in the dark at 22°C. For tests of vegetative-stage plants, seedlings grown for 10 days on plates under 16/8 h, 24/18°C, day/night cycle were treated at 38°C for 90 min followed by 2 h at 22°C and then 2 h at 45°C. Seedlings were photographed after 5 days.
SDS/PAGE and Western Blotting.
Etiolated seedlings (2.5 days old) of wild-type or hot1 mutant plants were pretreated at 38°C for 90 min, followed by 22°C for 2 h, and then total proteins were extracted in SDS sample buffer as described (27). Proteins were separated by SDS/PAGE on 7.5 or 12.5% acrylamide gels and processed for Western analysis (27). Protein blots were probed with rabbit antiserum against AtHsp101 (unpublished results) or AtHsp17.6 (27) or AtHsp22.0 (gift of K. Helm, Siena College, Laudonville, NY) at a dilution of 1:1000, and then with goat anti-rabbit horseradish peroxidase, and visualized by enhanced chemiluminescence (Amersham International).
Results
Identification of Four Loci Involved in Acquired Thermotolerance.
To identify mutants defective in acquired thermotolerance, we developed a quantitative assay for thermotolerance based on hypocotyl elongation in the dark. After 2.5 days of growth in the dark, seedlings are normally killed by a 2-h treatment at 45°C (Fig. 1; Col-O). However, if seedlings are first pretreated at 38°C for 90 min, a treatment which has no negative effect on growth, they will survive a subsequent 45°C stress, as determined by continued hypocotyl elongation and subsequent ability to grow normally in the light (Fig. 1; Col-O, and not shown).
Approximately 17,000 ethyl methanesulfonate-mutagenized M2 Arabidopsis seedlings, representing approximately 2,100 M1 parents, were screened to identify mutants that failed to show continued hypocotyl elongation after pretreatment at 38°C and subsequent 45°C stress (Materials and Methods). Importantly, for a significant percentage of mutants, the heat stress screening treatment was not lethal. That is, although the mutants failed to continue hypocotyl elongation directly after the stress treatment, many could be rescued after further cultivation at 22°C and grown to maturity. M3 seedlings of all putative mutants were retested to establish heritability of the phenotype. M3 seedlings were also tested to confirm that hypocotyl elongation was not affected by the 38°C pretreatment, but only by the subsequent 45°C stress. From this screen, four putative mutants were recovered.
The four mutants, which we have designated hot1, -2, -3, and -4, were backcrossed to the wild-type, parental Col-O ecotype for dominance and segregation testing. Eleven F1 seedlings for each mutant were tested for hypocotyl elongation after pretreatment and 45°C stress. All F1 seedlings behaved like wild type (not shown), establishing all four mutations as recessive. F2 seedlings from the backcross were subjected to the hypocotyl elongation test, and segregation of the mutant phenotype scored (Table 1). In all cases, segregation fits a 3:1 ratio of wild type/mutant, supporting the hypothesis that each mutation is caused by a single recessive allele.
Table 1.
Segregation of hot phenotype in F2
| No. of F2 seedlings
|
||
|---|---|---|
| WT | hot | |
| hot1 | 127 | 32 |
| hot2 | 106 | 35 |
| hot3 | 72 | 23 |
| hot4 | 160 | 52 |
All four mutants were also reciprocally crossed in all possible pairwise combinations, and resulting F1 seedlings tested for thermotolerance. F1 progeny of all crosses showed wild-type ability to grow in the hypocotyl elongation test (not shown). Therefore, the four hot mutations appear to represent distinct loci required for thermotolerance in Arabidopsis.
Phenotype of the hot Mutants.
A minimum of 18 F3 seedlings of hot1, hot2, and hot4, or M3 seedlings of hot3 were analyzed by the hypocotyl elongation test to obtain a quantitative description of this phenotype (Fig. 1 and Table 2). For all mutants, the 38°C pretreatment had no significant effect on hypocotyl elongation, but the subsequent 2-h, 45°C treatment severely curtailed their growth. The hot1, -2, and -3 mutants showed absolutely no growth, whereas the hot4 mutant showed slight growth after the 45°C treatment. hot4 growth could be completely arrested if the 45°C treatment was extended to 3 h, conditions that still allow growth of wild-type seedlings (not shown).
Table 2.
Hypocotyl elongation after heat shock
| 38°C | — | + | + |
|---|---|---|---|
| 45°C | — | — | + |
| Col-O | 12.6 ± 1.3 | 11.9 ± 1.6 | 6.2 ± 0.9 (49.2%) |
| hot1 | 12.6 ± 1.1 | 11.6 ± 1.1 | 0 ± 0 (0%) |
| hot2 | 4.2 ± 0.8 | 4.0 ± 0.9 | 0 ± 0 (0%) |
| hot3 | 11.7 ± 1.2 | 10.8 ± 0.8 | 0 ± 0 (0%) |
| hot4 | 11.6 ± 1.6 | 10.8 ± 2.0 | 1.0 ± 1.0 (8.6%) |
Measurement of hypocotyl growth after heat stress (in mm) was made on at least 18 seedlings from two independent assays. Each value represents the means ± SD. The numbers in parentheses indicate % elongation compared to that at 22°C.
Each of the mutants was also observed under optimal growth conditions for phenotypic alterations. hot1, -3, and -4 appear essentially wild type, with the exception of the wavy hypocotyl phenotype of hot4 (Fig. 1 and not shown). The hot2 mutation showed reduced hypocotyl length under all conditions compared with wild type, and appears semidwarf throughout its life cycle. It also produces excess lateral stems. To date these phenotypes have segregated with the thermotolerance defect, but further analysis will be required to confirm if the phenotypes are linked. All of the mutants were fertile and produced abundant seed with high germination rates. However, we have not yet determined if there are quantitative differences in fecundity or seed germination rates.
The hot1 Mutant Has a Point Mutation in the Hsp101 Gene.
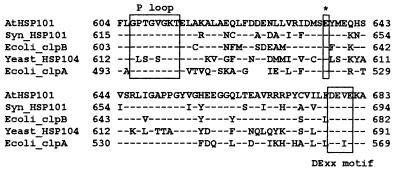
Mapping of the hot1 mutation to a chromosomal position was completed by standard molecular mapping techniques (Materials and Methods) by using 48 F2 outcrossed mutant progeny (Fig. 2). The hot1 mutation was located toward the bottom of chromosome 1, in the vicinity of the simple sequence-length polymorphic marker nga111, showing three recombinants with this marker out of 96 chromosomes scored. We noted that Hsp101, a gene we have found to be necessary for acquired thermotolerance (37), was within 500 kb of nga111 as determined from analysis of bacterial artificial chromosome end sequences on the corresponding bacterial artificial chromosome contig (http://genome-www.stanford.edu/Arabidopsis/). Thus, Hsp101 was a candidate gene for the hot1 mutation. To test this hypothesis, 6,609 bp of genomic DNA sequence encompassing the Hsp101 gene was determined for both wild type and the hot1 mutant. Within the region sequenced, only a single base pair change leading to a change in codon identity was found in the hot1 mutant. The codon for Glu-637 of Hsp101 was changed from GAA to AAA, resulting in substitution of a Lys residue (Fig. 2). Glu-637 is found within the second ATP-binding domain between the consensus P-loop and DExx motifs (4) (Fig. 3). Glu-637 is 100% conserved among evolutionarily diverse ClpB family members and the related E. coli ClpA. The conservation of this residue, and the fact that the mutation represented an extreme change in identity, from an acidic to a basic amino acid, was consistent with the conclusion that the Glu-637 → Lys mutation represented the lesion in the hot1 mutant.
Figure 3.
Glu-637 is conserved in divergent members of the Hsp100/ClpB family [Arabidopsis AtHsp101 (U13949), Synechococcus PCC7942 ClpB (U97124), E. coli ClpB (M29364), and S. cerevisiae Hsp104 (M67479)] and the ClpA family [E. coli ClpA (M31045)] of proteins. Relevant regions from the second ATP-binding domain of each protein are aligned. Residues identical to Hsp101 are indicated with dashes. A gap was introduced in E. coli ClpA to optimize the alignment.
The Wild-Type Hsp101 Gene Rescues the hot1 Phenotype.
To confirm that the hot1 mutation corresponded to a mutation in Hsp101, hot1 mutant plants were transformed with Arabidopsis genomic DNA as shown in Fig. 2, containing the wild-type Hsp101 gene controlled by its own promoter, or with vector alone. Seeds from three independent kanamycin-resistant plants transformed with Hsp101 or one kanamycin-resistant plant transformed with vector alone were analyzed for their thermotolerance phenotypes (Fig. 4). From the plants transformed with the Hsp101 gene, 14 of 24, 15 of 21, and 22 of 29 seedlings examined exhibited wild-type acquisition of thermotolerance for hypocotyl elongation (Fig. 4A), consistent with the expected dominance of the wild-type phenotype. The plant segregating 15 of 21 appeared to have two transgene insertions whereas the other two plants had single insertions as confirmed by kanamycin segregation testing. In contrast, all seedlings from the plant transformed with the vector alone showed only the hot1 mutant phenotype in the hypocotyl thermotolerance assay.
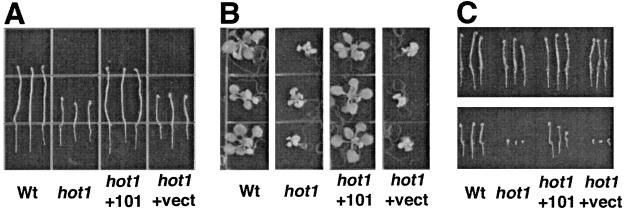
Figure 4.
Transformation of the hot1 mutant with an Hsp101 genomic clone reverts the thermotolerance phenotypes. Wt, untransformed Col-O; hot1, hot1 mutant; hot1 +101, mutant transformed with Hsp101 genomic clone; hot1 +vect, mutant transformed with vector only. (A) The 2.5-day-old, dark grown seedlings were treated at 38°C for 90 min followed by 2 h at 22°C and then 2 h at 45°C. Seedlings were returned to 22°C in the dark and photographed 2.5 days later. (B) The 10-day-old, light grown seedlings treated as in A and photographed 5 days later. (C) Seeds imbibed on plates at 4°C for 3 days were grown for 3 days at 22°C in the dark (Upper) or heated at 45°C for 2 h, returned to 22°C, and photographed after 3-days growth in the dark (Lower).
Because plants with reduced Hsp101 expression caused by antisense inhibition show reduced thermotolerance at later stages of growth (37), we also tested 10-day-old wild-type, mutant, and transformed plants for acquired thermotolerance (Fig. 4B). Similar to the hypocotyl assay, hot1 mutant seedlings fail to acquire thermotolerance at this stage of growth, but the expected proportion of progeny of plants transformed with the Hsp101 gene was restored to the wild-type phenotype.
It is well established that in comparison to seedlings or mature plants, germinating seeds (before radicle emergence) have a higher level of basal tolerance to high temperature and other stresses (28). In addition, Hsp101 accumulates during seed development and is present in dry seeds (unpublished results). We therefore tested the growth of hot1 seedlings treated directly after imbibition at 45°C for 2 h, a treatment that has little effect on germination and subsequent growth of the wild type. This stress treatment proved lethal to hot1 mutants; the radicle emerged, but no further growth was observed (Fig. 4C). This phenotype was also rescued by transformation with the Hsp101 gene.
Hsp101 and Other Hsps Accumulate at Normal Levels in hot1 Mutant Plants.
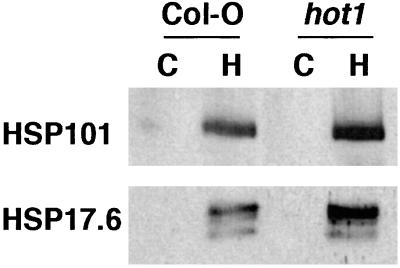
To determine how the hot1 mutation affected the expression of Hsp101 and other Hsps, total leaf proteins were analyzed by Western blotting with antibodies against Hsp101 and two small Hsps, Hsp17.6 (a cytosolic class I small Hsp; ref. 27) and Hsp22 (a small Hsp localized to the endoplasmic reticulum; ref. 29). The hot1 mutant accumulated wild-type levels of all three of these Hsps after a heat stress (Fig. 5, and not shown). Thus, the hot1 mutant does not appear to be generally impaired in the heat stress response, and the defect in Hsp101 in hot1 does not appear to destabilize the protein.
Figure 5.
Hsp101 and small Hsps accumulate normally in mutant seedlings after heat stress. Total proteins from 2.5-day-old dark grown seedlings were isolated either before (C) or after treatment at 38°C for 90 min, followed by 2 h at 22°C (H). Proteins were analyzed by Western blotting with either Hsp101 or Hsp17.6 antibodies.
Discussion
By using a screen designed to identify mutants defective in the ability to acquire tolerance to high temperature, we have defined four genetic loci required for this process in Arabidopsis. We further showed that one of these loci, hot1, encodes the Hsp101 gene, the only gene established to be required for acquired thermotolerance in plants (37). This result provides direct verification that the screen is targeting genes critical to the temperature adaptation process. Identification of the hot mutations represents the first successful attempt toward a genetic dissection of thermotolerance mechanisms in a higher eukaryote. In addition, the hot1 mutation is the first defined mutation in any Hsp in plants.
The hot mutations described here were all isolated from ethyl methanesulfonate-mutagenized material, suggesting that they arise from point mutations, as seen for hot1. Although point mutations can frequently lead to temperature sensitivity of proteins, it seems unlikely that our screen is simply identifying temperature-sensitive proteins involved in housekeeping functions. First, as mentioned above, we have already established that Hsp101 is required for thermotolerance from antisense experiments in Arabidopsis (37). In the antisense experiments, the loss-of-function phenotype arises from near absence of wild-type protein. Second, the defect in the hot mutants is observed only after the 45°C treatment; they all show normal growth after the 38°C pretreatment. This phenotype indicates that the corresponding gene products would be temperature sensitive only above 38°C. Whereas this is possible, the temperature sensitivity must also have the property that only a short 45°C treatment (2 h) results in inactivation that is not reversed during 2.5–5 days of growth at normal temperatures. We are not aware of other temperature-sensitive mutations with such properties. Altogether, the data argue that the hot loci represent essential components required specifically for adaptation to high temperature.
It is of interest to consider the nature of the defect in Hsp101 in the hot1 mutant. We observed that the mutant protein accumulates normally. Thus, it is unlikely that the mutation severely compromises the structure of the protein, because that would be predicted to lead to degradation of the mutant protein. To understand the position of the Glu-637 → Lys mutation relative to the conserved motifs of the ATP-binding domain, the HSP101 sequence was aligned with the ATP-binding domain from other members of the AAA+ family of ATPases (4) and then superimposed on the known three-dimensional structure of the δ′ subunit of E. coli DNA polymerase III (ref. 30 and not shown). Based on this analysis, the hot1 mutation would be positioned toward the end of the β2 strand directly adjacent to the DExx motif, in a position that could interfere with proper ATP binding and hydrolysis. It is already established from mutational analysis of the P-loop that both ATP-binding sites are required for the thermotolerance function of Hsp104 in yeast (31). We have found no reports characterizing any mutations outside of the P-loop in any Hsp100/ClpB protein. Expression and purification of the mutant protein can be used to test directly for alterations in its ATPase activity. The same mutation can also be introduced into yeast Hsp104 to determine its effect on acquired thermotolerance in vivo in that organism.
The fact that the hot1 mutation is recessive, despite high levels of mutant protein accumulation, indicates that the mutant Hsp101 does not interfere with function of the wild-type protein. Hsp100/ClpB proteins are hexamers in the native state, and presumably the oligomeric structure is necessary for function (3). This means either that the mutant protein does not stably coassemble with wild-type protein, or if it does coassemble, wild-type subunits still function in the context of mutant neighbor subunits, or enough oligomeric wild-type protein is available even in the presence of mixed-subunit oligomers.
We do not know if the hot1 mutation exhibits a null phenotype for Hsp101; it may retain partial activity, sufficient for essential functions during normal growth, but not for the development of thermotolerance. Besides being expressed during heat stress (32), Hsp101 is also expressed during seed development (unpublished results), and homologous proteins are regulated by other stresses in other plant species (33). In addition to an essential function in conferring basal thermotolerance to seeds, it is possible that Hsp101 is important for optimal seed development or other functions that remain to be determined. If Hsp101 functions under optimal growth conditions in addition to during stress, these functions may be partially redundant with the function of other homologous genes, explaining the relative normal phenotype of hot1 plants in the absence of stress. The Arabidopsis genome project has identified two Hsp101 homologues, Hsp92.7 (D71409) and Hsp98.7 (CAA20530), with 74 and 65% similarity to Hsp101, respectively. Both genes are predicted to encode cytosolic proteins. Hsp92.7 is also expressed during heat stress, but is not required for thermotolerance (Hong, S.-W. & Vierling, E., unpublished results). No information is available on expression of Hsp98.7. How these genes may compensate for the loss of Hsp101 during normal growth or under other stress conditions remains to be determined.
Although overexpression of Hsp101 confers some increased thermotolerance to Arabidopsis (37), the fact that we have identified other loci, unlinked to hot1 but with a related phenotype, indicates other factors also contribute to thermotolerance. Thus, Hsp101 is clearly necessary, but not sufficient for acquired thermotolerance in plants. The other hot mutations might represent other Hsps (21) or genes involved in regulation of Hsps, such as heat shock transcription factor genes (34). In addition to causing protein denaturation, a problem that appears to be ameliorated by Hsps, high temperature also alters membrane fluidity, can disrupt the overall balance of metabolic processes (35), and interferes with protein translation (36). Cloning of the other hot loci will help answer the question of what additional processes make a major contribution to thermotolerance.
It will be of interest to examine all the hot mutants for their phenotypes after different acute and chronic high-temperature treatments during various life stages, as well as during other stress treatments. How acquired thermotolerance might relate to homeostasis of cell function under fluctuating environmental temperatures is unknown, and should be testable with the hot mutants. In addition, suppressors of the hot1 mutation should be easily isolated by using a simple screen for wild-type growth of heat-treated mutant seeds. Thus, the hot1 mutation, as well as other alleles that may be identified at this locus, afford a unique opportunity for genetic dissection of the structure and mechanism of action of an Hsp in higher plants.
Acknowledgments
We thank Chris Borchert and Sarah Ryan for untiring help in the mutant screen, Dr. K. Helm for the gift of Hsp22 antibodies, and Drs. K. Giese and F. Tax for critical reading of the manuscript. Support was provided by grants from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (96–351003232), the Department of Energy Energy Biosciences Program (95ER20208), and the University of Arizona Experiment Station to E.V.
Abbreviation
- Hsp
heat shock protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF218796).
References
- 1.Lindquist S. Annu Rev Biochem. 1986;45:39–72. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 2.Trent J D, Gabrielsen M, Jensen B, Neuhard J, Olsen J. J Bacteriol. 1994;176:6148–6152. doi: 10.1128/jb.176.19.6148-6152.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schirmer E C, Glover J R, Singer M A, Lindquist S. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 4.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 5.Sanchez Y, Lindquist S L. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 6.Eriksson M J, Clarke A K. J Bacteriol. 1996;178:4839–4846. doi: 10.1128/jb.178.16.4839-4846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsell D A, Kowal A S, Singer M A, Lindquist S. Nature (London) 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 8.Glover J R, Lindquist S. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 9.Zolkiewski M. J Biol Chem. 1999;274:28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]
- 10.Motohashi K, Watanabe Y, Yohda M, Yoshida M. Proc Natl Acad Sci USA. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin D-Y, Matsumoto K, Iida H, Uno I, Ishikawa T. Mol Cell Biol. 1987;7:244–250. doi: 10.1128/mcb.7.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piper P W. FEMS Microbiol Lett. 1993;11:339–355. doi: 10.1111/j.1574-6976.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 13.Fung P A, Gaertig J, Gorovsky M A, Hallberg R L. Science. 1995;268:1036–1039. doi: 10.1126/science.7754381. [DOI] [PubMed] [Google Scholar]
- 14.Plesofsky-Vig N, Brambl R. Proc Natl Acad Sci USA. 1995;92:5032–5036. doi: 10.1073/pnas.92.11.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S, Prochaska D J, Fang F, Barnum S R. Curr Microbiol. 1998;37:403–407. doi: 10.1007/s002849900400. [DOI] [PubMed] [Google Scholar]
- 16.Petko L, Lindquist S. Cell. 1985;45:885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- 17.Vierling E. Acta Physiol Plantarum. 1997;19:539–547. [Google Scholar]
- 18.Parsell D A, Taulien J, Lindquist S. Philos Trans R Soc London B. 1993;339:279–286. doi: 10.1098/rstb.1993.0026. [DOI] [PubMed] [Google Scholar]
- 19.Lindquist S. Curr Opin Genet Dev. 1992;2:748–755. doi: 10.1016/s0959-437x(05)80135-2. [DOI] [PubMed] [Google Scholar]
- 20.Radin J W, Lu Z, Percy R G, Zeiger E. Proc Natl Acad Sci USA. 1994;91:7217–7221. doi: 10.1073/pnas.91.15.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vierling E. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:579–620. [Google Scholar]
- 22.Haughn G W, Somerville C. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- 23.Klimyuk V I, Carroll B J, Thomas C M, Jones J D G. Plant J. 1993;3:493–494. doi: 10.1111/j.1365-313x.1993.tb00169.x. [DOI] [PubMed] [Google Scholar]
- 24.Bell C J, Ecker J R. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 25.Frisch D A, Harris-Haller L W, Yokubaitis N T, Thomas T L, Hardin S H, Hall T H. Plant Mol Biol. 1995;27:405–409. doi: 10.1007/BF00020193. [DOI] [PubMed] [Google Scholar]
- 26.Bechtold N, Ellis J, Pelletier G. C R Acad Sci Ser III. 1993;316:1194–1199. [Google Scholar]
- 27.Wehmeyer N, Hernandez L D, Finkelstein R R, Vierling E. Plant Physiol. 1996;112:747–757. doi: 10.1104/pp.112.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bewley J D, Black M. Seeds: Physiology of Development and Germination. New York: Plenum; 1985. pp. 1–367. [Google Scholar]
- 29.Helm K W, Schmeits J, Vierling E. Plant Physiol. 1995;107:287–288. doi: 10.1104/pp.107.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guenther B, Onrust R, Sali A, O'Donnell M, Kuriyan J. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 31.Parsell D A, Sanchez Y, Stitzel J D, Lindquist S. Nature (London) 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- 32.Schirmer E C, Lindquist S, Vierling E. Plant Cell. 1994;6:1899–1909. doi: 10.1105/tpc.6.12.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singla S L, Pareek A, Grover A. Plant Sci (Limerick, Irel) 1997;125:211–219. [Google Scholar]
- 34.Nover L, Scharf K-D, Gagliard D, Vergne P, Czarnecke-Verner E, Gurley W B. Cell Stress Chaperones. 1996;1:215–223. doi: 10.1379/1466-1268(1996)001<0215:thwcap>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dewey W C. Radiat Res. 1989;120:191–204. [PubMed] [Google Scholar]
- 36.McCormick W, Penman S. J Mol Biol. 1969;39:315–333. doi: 10.1016/0022-2836(69)90320-9. [DOI] [PubMed] [Google Scholar]
- 37.Queitsch, C., Hong, S.-W., Vierling, E. & Lindquist, S. (2000) Plant Cell, in press. [DOI] [PMC free article] [PubMed]