Abstract
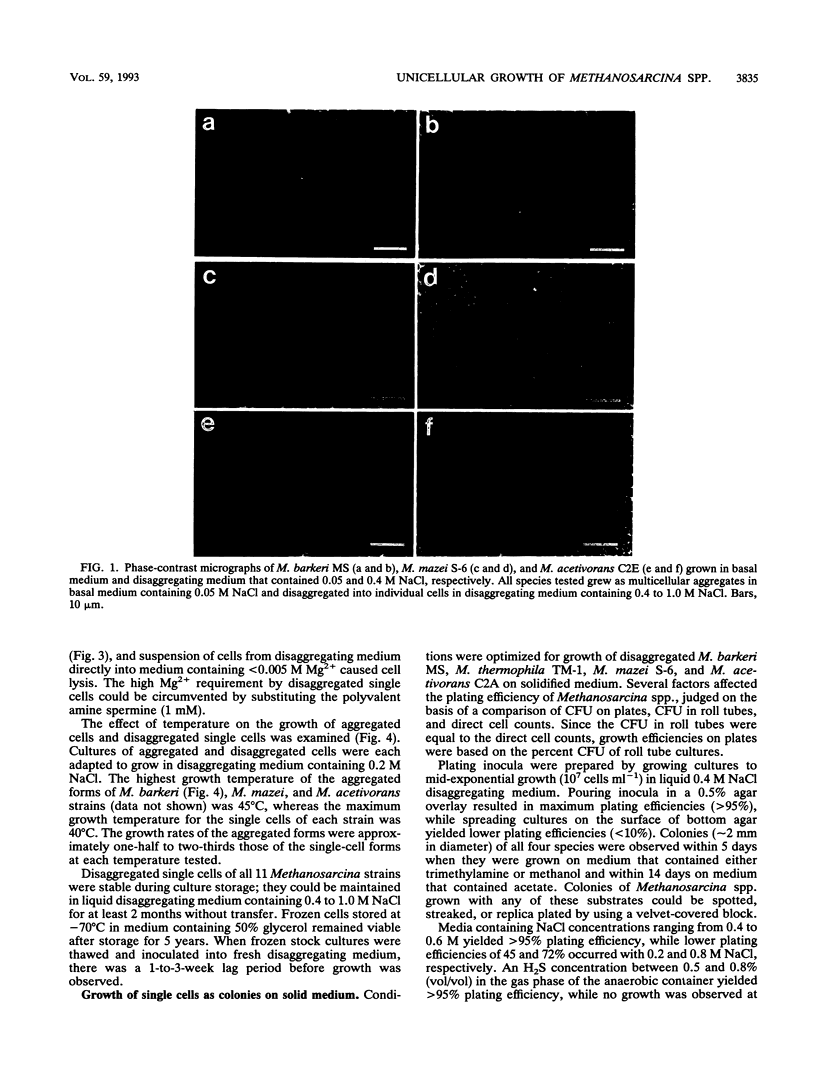
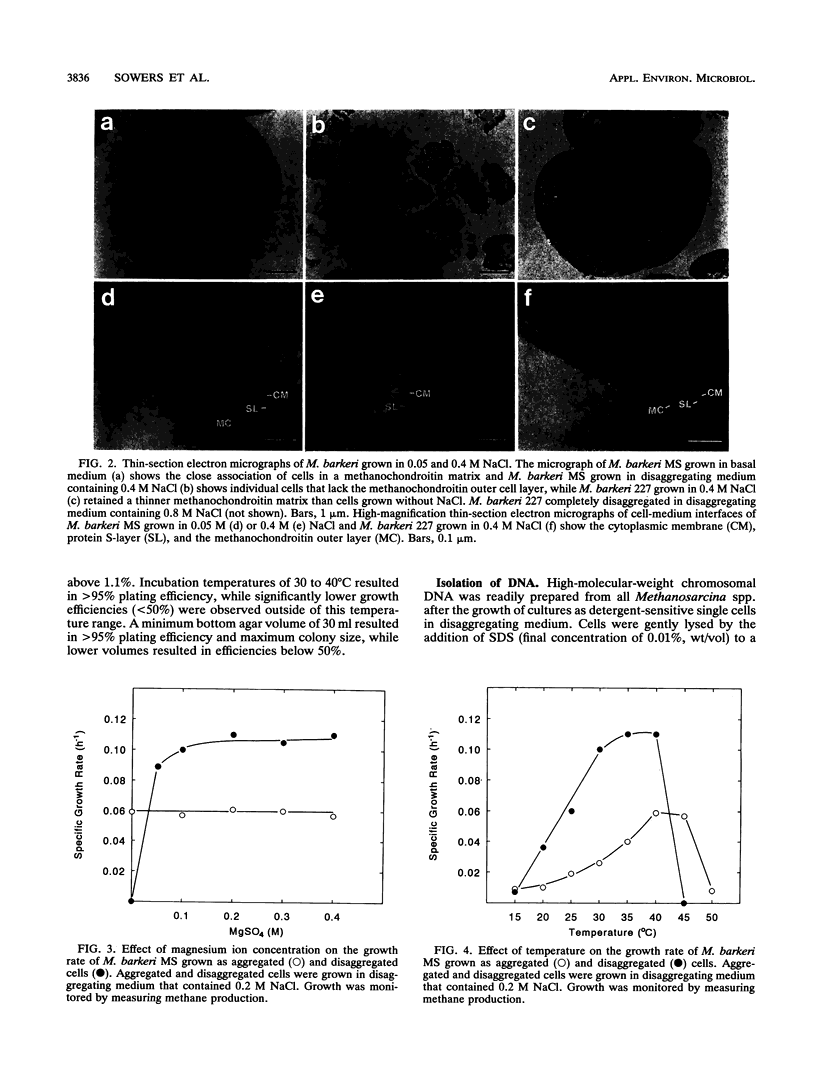
The effect of medium osmolarity on the morphology and growth of Methanosarcina barkeri, Methanosarcina thermophila, Methanosarcina mazei, Methanosarcina vacuolata, and Methanosarcina acetivorans was examined. Each strain was adapted for growth in NaCl concentrations ranging from 0.05 to 1.0 M. Methanosarcina spp. isolated from both marine and nonmarine sources exhibited similar growth characteristics at all NaCl concentrations tested, demonstrating that these species are capable of adapting to a similar range of medium osmolarities. Concomitant with the adaptation in 0.4 to 1.0 M NaCl, all strains disaggregated and grew as single cells rather than in the characteristic multicellular aggregates. Aggregated cells had a methanochondroitin outer layer, while disaggregated single cells lacked the outer layer but retained the protein S-layer adjacent to the cell membrane. Synthesis of glucuronic acid, a major component of methanochondroitin, was reduced 20-fold in the single-cell form of M. barkeri when compared with synthesis in aggregated cells. Strains with the methanochondroitin outer cell layer exhibited enhanced stability at low (<0.2 M NaCl) osmolarity and grew at higher temperatures. Disaggregated cells could be converted back to aggregated cells by gradually readapting cultures to lower NaCl (<0.2 M) and Mg2+ (<0.005 M) concentrations. Disaggregated Methanosarcina spp. could also be colonized and replica plated with greater than 95% recovery rates on solidified agar basal medium that contained 0.4 to 0.6 M NaCl and either trimethylamine, methanol, or acetate as the substrate. The ability to disaggregate and grow Methanosarcina spp. as viable, detergent-sensitive, single cells on agar medium makes these species amenable to mutant selection and screening for genetic studies and enables cells to be gently lysed for the isolation of intact genetic material.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards T., McBride B. C. New method for the isolation and identification of methanogenic bacteria. Appl Microbiol. 1975 Apr;29(4):540–545. doi: 10.1128/am.29.4.540-545.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. E. Spontaneous Disaggregation of Methanosarcina mazei S-6 and Its Use in the Development of Genetic Techniques for Methanosarcina spp. Appl Environ Microbiol. 1987 Oct;53(10):2500–2504. doi: 10.1128/aem.53.10.2500-2504.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. J., Nagle D. P., Jr, Whitman W. B. Methanogens and the diversity of archaebacteria. Microbiol Rev. 1987 Mar;51(1):135–177. doi: 10.1128/mr.51.1.135-177.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. J., Whitman W. B., Fields R. D., Wolfe R. S. Growth and plating efficiency of methanococci on agar media. Appl Environ Microbiol. 1983 Jul;46(1):220–226. doi: 10.1128/aem.46.1.220-226.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O., Hippe H. Lack of peptidoglycan in the cell walls of Methanosarcina barkeri. Arch Microbiol. 1977 May 13;113(1-2):57–60. doi: 10.1007/BF00428580. [DOI] [PubMed] [Google Scholar]
- Liu Y., Boone D. R., Sleat R., Mah R. A. Methanosarcina mazei LYC, a New Methanogenic Isolate Which Produces a Disaggregating Enzyme. Appl Environ Microbiol. 1985 Mar;49(3):608–613. doi: 10.1128/aem.49.3.608-613.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Klug M. J. Intermediary metabolism of organic matter in the sediments of a eutrophic lake. Appl Environ Microbiol. 1982 Mar;43(3):552–560. doi: 10.1128/aem.43.3.552-560.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundie L. L., Jr, Ferry J. G. Activation of acetate by Methanosarcina thermophila. Purification and characterization of phosphotransacetylase. J Biol Chem. 1989 Nov 5;264(31):18392–18396. [PubMed] [Google Scholar]
- Sowers K. R., Baron S. F., Ferry J. G. Methanosarcina acetivorans sp. nov., an Acetotrophic Methane-Producing Bacterium Isolated from Marine Sediments. Appl Environ Microbiol. 1984 May;47(5):971–978. doi: 10.1128/aem.47.5.971-978.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Ferry J. G. Isolation and Characterization of a Methylotrophic Marine Methanogen, Methanococcoides methylutens gen. nov., sp. nov. Appl Environ Microbiol. 1983 Feb;45(2):684–690. doi: 10.1128/aem.45.2.684-690.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Gunsalus R. P. Adaptation for growth at various saline concentrations by the archaebacterium Methanosarcina thermophila. J Bacteriol. 1988 Feb;170(2):998–1002. doi: 10.1128/jb.170.2.998-1002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Gunsalus R. P. Plasmid DNA from the acetotrophic methanogen Methanosarcina acetivorans. J Bacteriol. 1988 Oct;170(10):4979–4982. doi: 10.1128/jb.170.10.4979-4982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Robertson D. E., Noll D., Gunsalus R. P., Roberts M. F. N epsilon-acetyl-beta-lysine: an osmolyte synthesized by methanogenic archaebacteria. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9083–9087. doi: 10.1073/pnas.87.23.9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOLIN E. A., WOLIN M. J., WOLFE R. S. FORMATION OF METHANE BY BACTERIAL EXTRACTS. J Biol Chem. 1963 Aug;238:2882–2886. [PubMed] [Google Scholar]
- Weiss R. L. Subunit cell wall of Sulfolobus acidocaldarius. J Bacteriol. 1974 Apr;118(1):275–284. doi: 10.1128/jb.118.1.275-284.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrell V. E., Nagle D. P., Jr, McCarthy D., Eisenbraun A. Genetic transformation system in the archaebacterium Methanobacterium thermoautotrophicum Marburg. J Bacteriol. 1988 Feb;170(2):653–656. doi: 10.1128/jb.170.2.653-656.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun L. Y., Mah R. A., Boone D. R. Isolation and characterization of disaggregatase from Methanosarcina mazei LYC. Appl Environ Microbiol. 1990 Dec;56(12):3693–3698. doi: 10.1128/aem.56.12.3693-3698.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun L., Boone D. R., Mah R. A. Control of the Life Cycle of Methanosarcina mazei S-6 by Manipulation of Growth Conditions. Appl Environ Microbiol. 1988 Aug;54(8):2064–2068. doi: 10.1128/aem.54.8.2064-2068.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]