Abstract
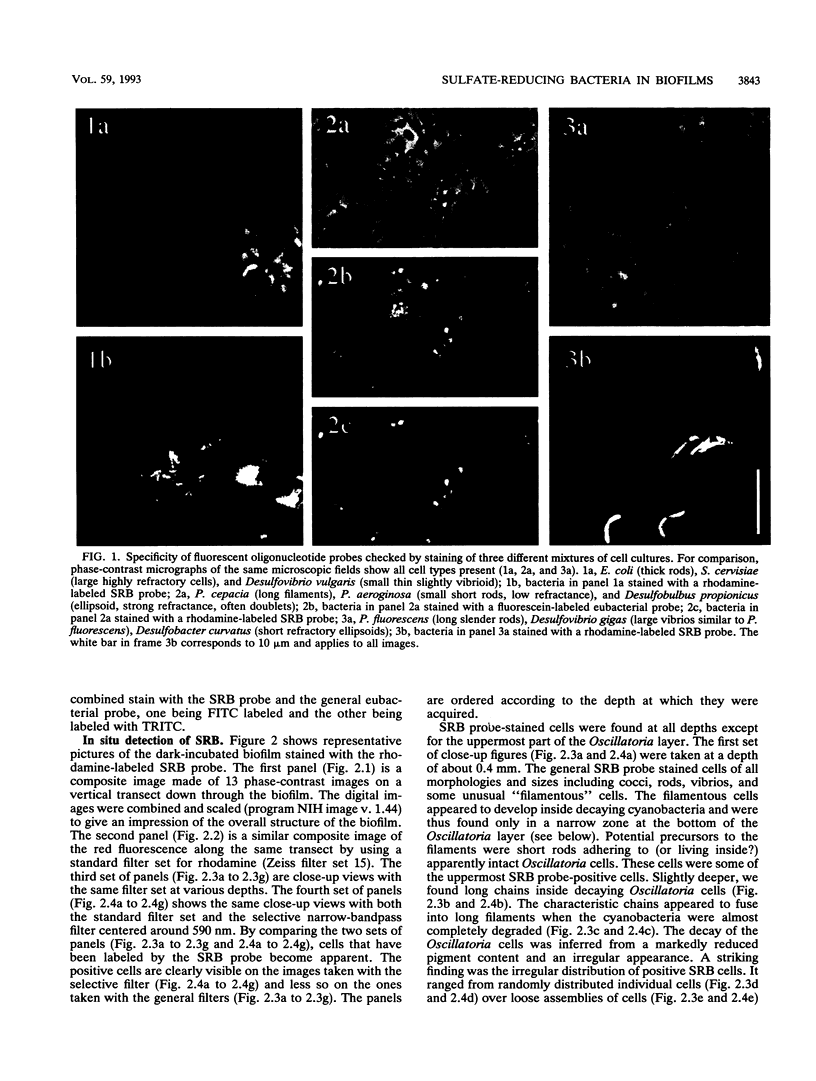
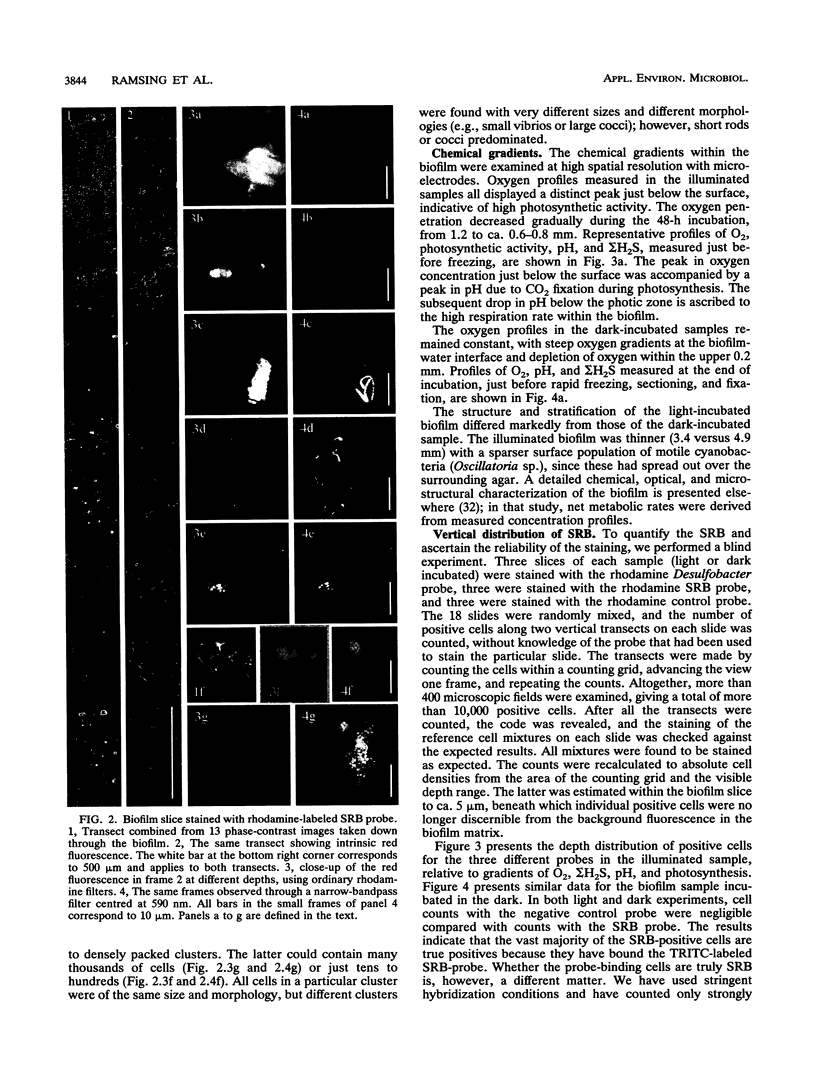
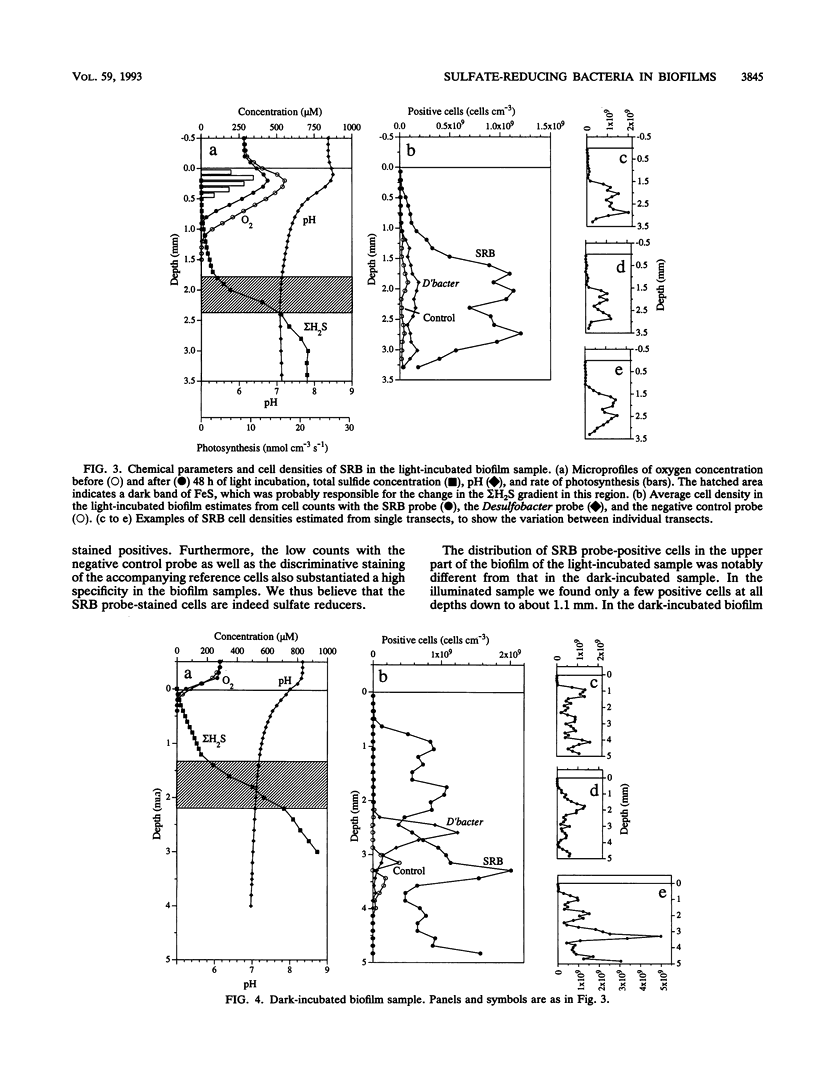
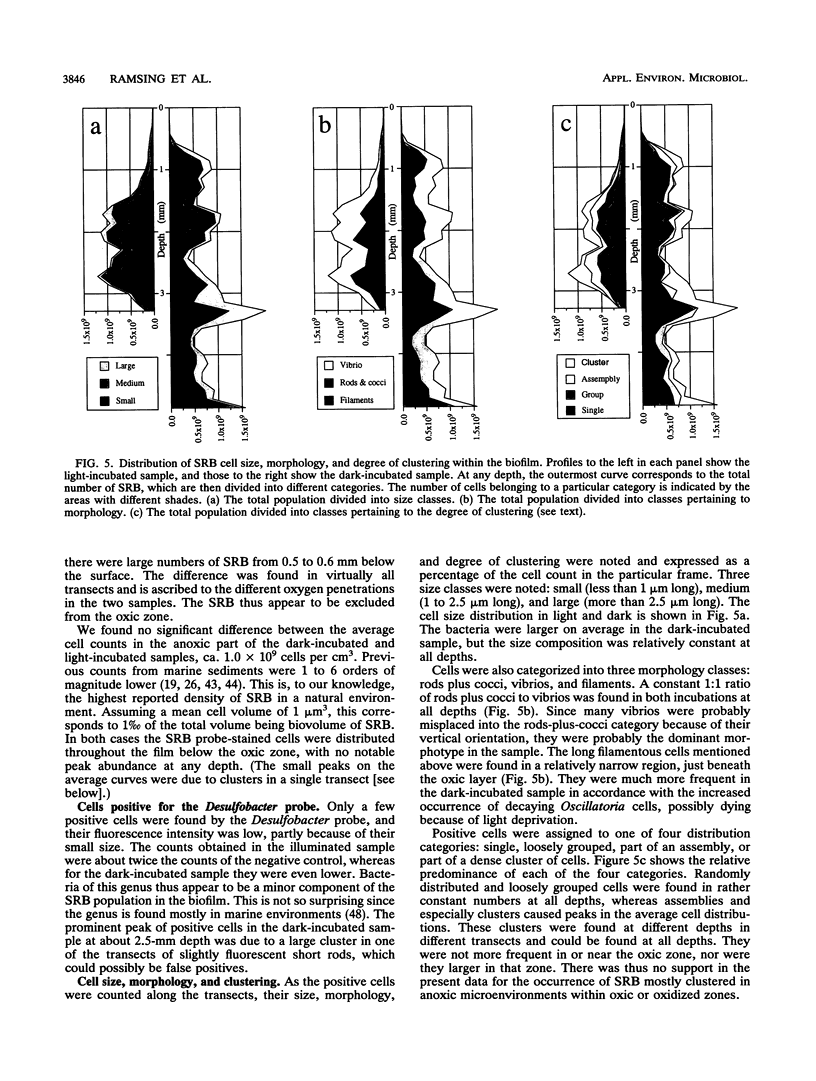
The vertical distribution of sulfate-reducing bacteria (SRB) in photosynthetic biofilms from the trickling filter of a sewage treatment plant was investigated with oligonucleotide probes binding to 16S rRNA. To demonstrate the effect of daylight and photosynthesis and thereby of increased oxygen penetration, we incubated two 4-mm-thick biofilm samples in darkness or exposed to light at natural intensity. Gradients of O2, H2S, and pH were examined with microelectrodes during incubation. The samples were subsequently frozen with liquid nitrogen and sliced on a cryomicrotome in 20-microns vertical slices. Fluorescent-dye-conjugated oligonucleotides were used as "phylogenetic" probes to identify single cells in the slices. Oligonucleotide sequences were selected which were complementary to short sequence elements (16 to 20 nucleotides) within the 16S rRNA of sulfate-reducing bacteria. The probes were labeled with fluorescein or rhodamine derivatives for subsequent visualization by epifluorescence microscopy. Five probes were synthesized for eukaryotes, eubacteria, SRB (including most species of the delta group of purple bacteria), Desulfobacter spp., and a nonhybridizing control. The SRB were unevenly distributed in the biofilm, being present in all states from single scattered cells to dense clusters of several thousand cells. To quantify the vertical distribution of SRB, we counted cells along vertical transects through the biofilm. This was done in a blind experiment to ascertain the reliability of the staining. A negative correlation between the vertical distribution of positively stained SRB cells and the measured O2 profiles was found. The distribution differed in light- and dark-incubated samples presumably because of the different extensions of the oxic surface layer. In both cases the SRB were largely restricted to anoxic layers.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agard D. A. Optical sectioning microscopy: cellular architecture in three dimensions. Annu Rev Biophys Bioeng. 1984;13:191–219. doi: 10.1146/annurev.bb.13.060184.001203. [DOI] [PubMed] [Google Scholar]
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990 Jun;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Stromley J., Devereux R., Key R., Stahl D. A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992 Feb;58(2):614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Canfield D. E., Des Marais D. J. Aerobic sulfate reduction in microbial mats. Science. 1991 Mar 22;251:1471–1473. doi: 10.1126/science.11538266. [DOI] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Devereux R., Delaney M., Widdel F., Stahl D. A. Natural relationships among sulfate-reducing eubacteria. J Bacteriol. 1989 Dec;171(12):6689–6695. doi: 10.1128/jb.171.12.6689-6695.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T., Ramsing N. B. Identification of sulphate-reducing ectosymbiotic bacteria from anaerobic ciliates using 16S rRNA binding oligonucleotide probes. Arch Microbiol. 1992;158(6):394–397. doi: 10.1007/BF00276298. [DOI] [PubMed] [Google Scholar]
- Fründ C., Cohen Y. Diurnal Cycles of Sulfate Reduction under Oxic Conditions in Cyanobacterial Mats. Appl Environ Microbiol. 1992 Jan;58(1):70–77. doi: 10.1128/aem.58.1.70-77.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks R. E., Amann R. I., Stahl D. A. Dual staining of natural bacterioplankton with 4',6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992 Jul;58(7):2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurtshuk R. J., Blick M., Bresser J., Fox G. E., Jurtshuk P., Jr Rapid in situ hybridization technique using 16S rRNA segments for detecting and differentiating the closely related gram-positive organisms Bacillus polymyxa and Bacillus macerans. Appl Environ Microbiol. 1992 Aug;58(8):2571–2578. doi: 10.1128/aem.58.8.2571-2578.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen B. B., Bak F. Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (kattegat, denmark). Appl Environ Microbiol. 1991 Mar;57(3):847–856. doi: 10.1128/aem.57.3.847-856.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl M., Jørgensen B. B. Microsensor measurements of sulfate reduction and sulfide oxidation in compact microbial communities of aerobic biofilms. Appl Environ Microbiol. 1992 Apr;58(4):1164–1174. doi: 10.1128/aem.58.4.1164-1174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laanbroek H. J., Pfennig N. Oxidation of short-chain fatty acids by sulfate-reducing bacteria in freshwater and in marine sediments. Arch Microbiol. 1981 Jan;128(3):330–335. doi: 10.1007/BF00422540. [DOI] [PubMed] [Google Scholar]
- Lathe R. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J Mol Biol. 1985 May 5;183(1):1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Ramsing N. B., Jovin T. M. Parallel stranded duplex DNA. Nucleic Acids Res. 1988 Jul 25;16(14A):6659–6676. doi: 10.1093/nar/16.14.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsing N. B., Rippe K., Jovin T. M. Helix-coil transition of parallel-stranded DNA. Thermodynamics of hairpin and linear duplex oligonucleotides. Biochemistry. 1989 Nov 28;28(24):9528–9535. doi: 10.1021/bi00450a042. [DOI] [PubMed] [Google Scholar]
- Rehnstam A. S., Norqvist A., Wolf-Watz H., Hagström A. Identification of Vibrio anguillarum in fish by using partial 16S rRNA sequences and a specific 16S rRNA oligonucleotide probe. Appl Environ Microbiol. 1989 Aug;55(8):1907–1910. doi: 10.1128/aem.55.8.1907-1910.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Bratina B. J., Tsuji K., Hanson R. S. Use of oligodeoxynucleotide signature probes for identification of physiological groups of methylotrophic bacteria. Appl Environ Microbiol. 1990 Sep;56(9):2858–2865. doi: 10.1128/aem.56.9.2858-2865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viles C. L., Sieracki M. E. Measurement of marine picoplankton cell size by using a cooled, charge-coupled device camera with image-analyzed fluorescence microscopy. Appl Environ Microbiol. 1992 Feb;58(2):584–592. doi: 10.1128/aem.58.2.584-592.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. T., van Gemerden H. Production and consumption of dimethylsulfoniopropionate in marine microbial mats. Appl Environ Microbiol. 1991 Nov;57(11):3237–3242. doi: 10.1128/aem.57.11.3237-3242.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Amann R., Lemmer H., Schleifer K. H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993 May;59(5):1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. M., Weller R., Bateson M. M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990 May 3;345(6270):63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- Zarda B., Amann R., Wallner G., Schleifer K. H. Identification of single bacterial cells using digoxigenin-labelled, rRNA-targeted oligonucleotides. J Gen Microbiol. 1991 Dec;137(12):2823–2830. doi: 10.1099/00221287-137-12-2823. [DOI] [PubMed] [Google Scholar]