Abstract
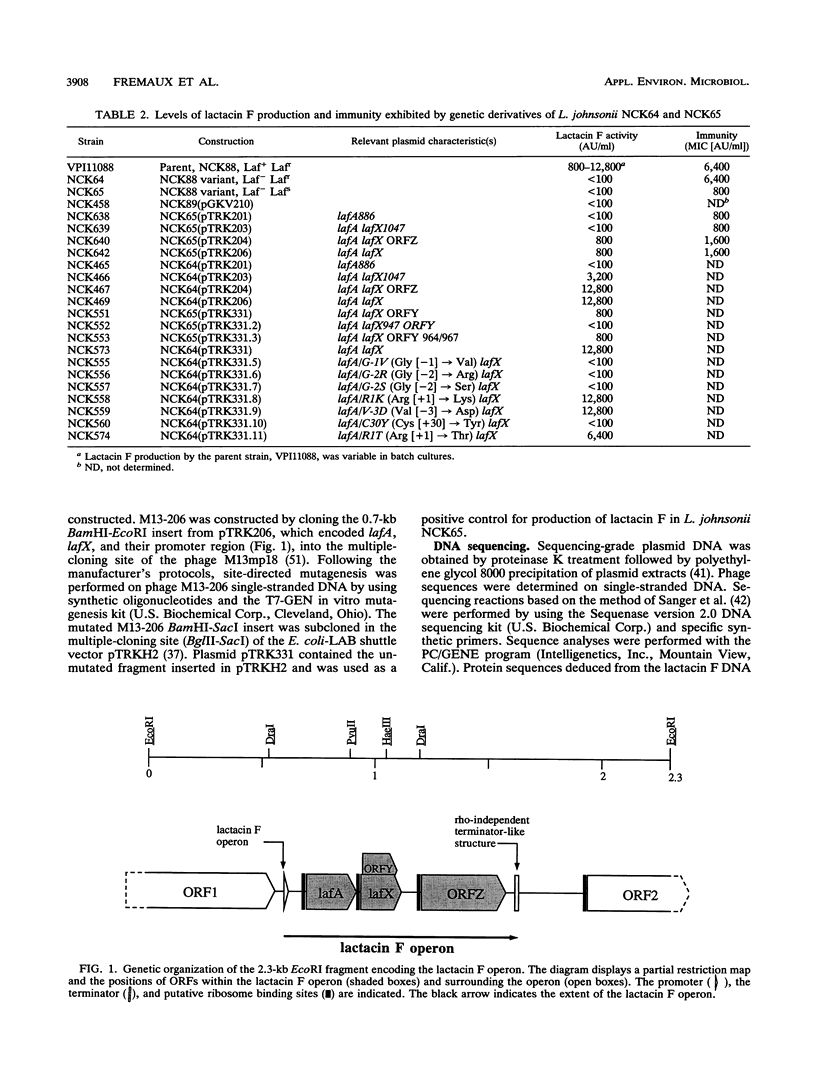
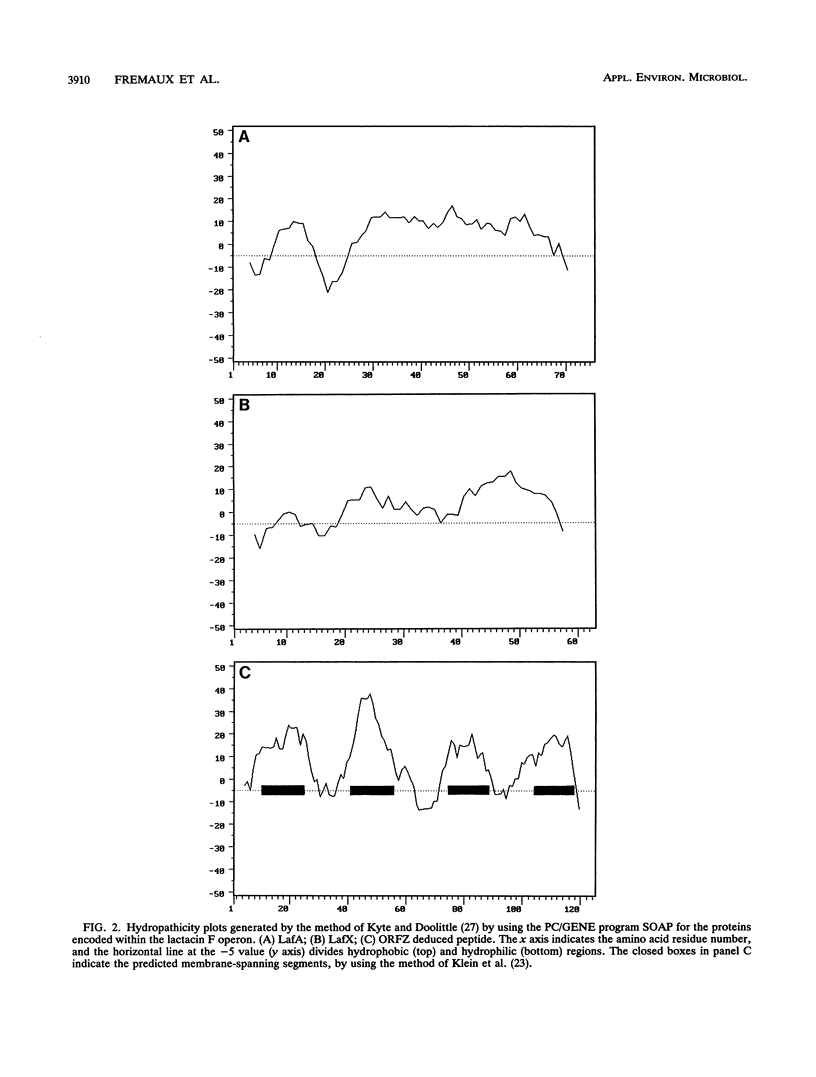
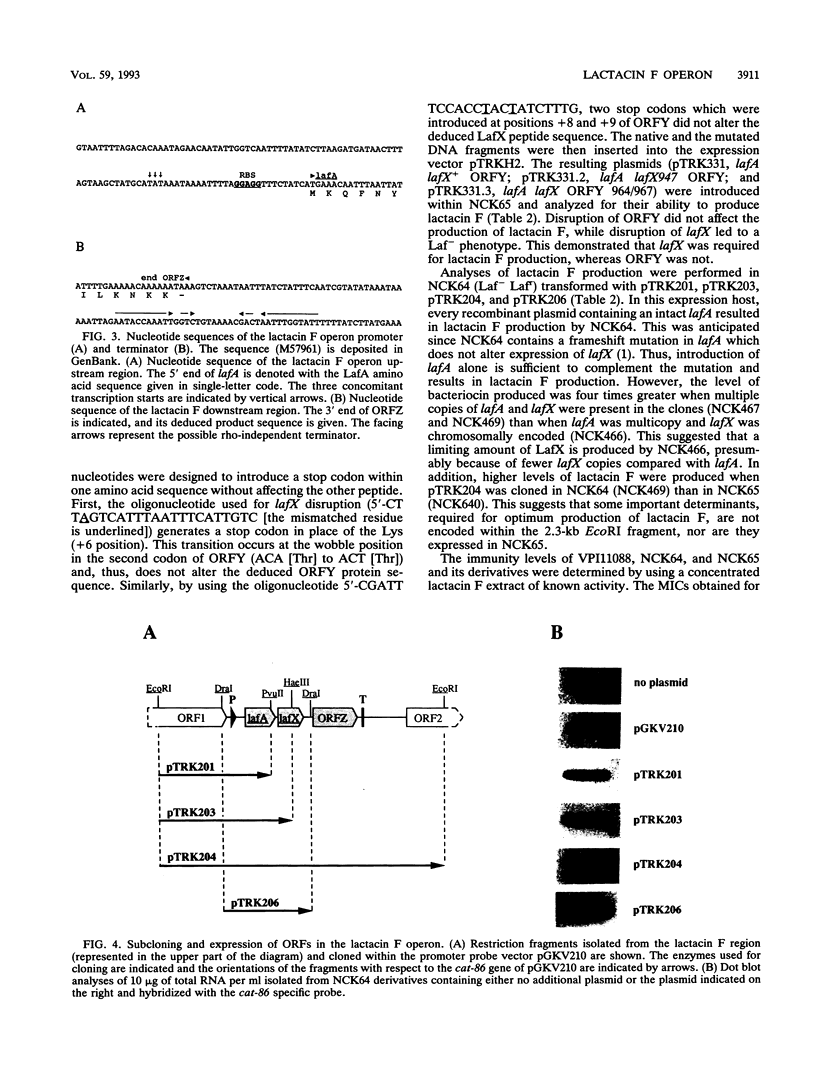
Lactacin F is a nonlantibiotic, heat-stable, peptide bacteriocin produced by Lactobacillus johnsonii VPI11088. Molecular analysis of the lactacin F DNA region characterized a small operon that codes for three open reading frames, designated lafA, lafX, and ORFZ. The peptide encoded by lafA, the lactacin F structural gene, was compared with various peptide bacteriocins from lactic acid bacteria, and similarities were identified in the amino and carboxy termini of the propeptides. Site-directed mutagenesis of the LafA precursor at the two glycine residues in positions -1 and -2 defined an essential motif for processing of mature lactacin F. The involvement of the peptides encoded by lafX and ORFZ in bacteriocin expression was investigated by subcloning various fragments from the lactacin F region into the shuttle vector pGKV210. In addition to lafA, expression of lafX is essential to lactacin F activity. The lactacin F operon resembles the genetic organization of lactococcin M. Although no function has been assigned to ORFZ by genetic analysis, both peptide Z and the lactococcin M immunity protein are predicted to be integral membrane proteins with four putative transmembrane segments. Lactacin F activity, defined by bactericidal action on Lactobacillus delbrueckii, is dependent on the expression of two genes, lafA and lafX.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barefoot S. F., Klaenhammer T. R. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl Environ Microbiol. 1983 Jun;45(6):1808–1815. doi: 10.1128/aem.45.6.1808-1815.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke G., Gutowski-Eckel Z., Hammelmann M., Entian K. D. Biosynthesis of the lantibiotic nisin: genomic organization and membrane localization of the NisB protein. Appl Environ Microbiol. 1992 Nov;58(11):3730–3743. doi: 10.1128/aem.58.11.3730-3743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T., Benno Y., Yaeshima T., Mitsuoka T. Taxonomic study of the Lactobacillus acidophilus group, with recognition of Lactobacillus gallinarum sp. nov. and Lactobacillus johnsonii sp. nov. and synonymy of Lactobacillus acidophilus group A3 (Johnson et al. 1980) with the type strain of Lactobacillus amylovorus (Nakamura 1981). Int J Syst Bacteriol. 1992 Jul;42(3):487–491. doi: 10.1099/00207713-42-3-487. [DOI] [PubMed] [Google Scholar]
- Gao F. H., Abee T., Konings W. N. Mechanism of action of the peptide antibiotic nisin in liposomes and cytochrome c oxidase-containing proteoliposomes. Appl Environ Microbiol. 1991 Aug;57(8):2164–2170. doi: 10.1128/aem.57.8.2164-2170.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Geis A., Singh J., Teuber M. Potential of lactic streptococci to produce bacteriocin. Appl Environ Microbiol. 1983 Jan;45(1):205–211. doi: 10.1128/aem.45.1.205-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves M. C., Rabinowitz J. C. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. Evidence for "extended" promoter elements in gram-positive organisms. J Biol Chem. 1986 Aug 25;261(24):11409–11415. [PubMed] [Google Scholar]
- Hastings J. W., Sailer M., Johnson K., Roy K. L., Vederas J. C., Stiles M. E. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J Bacteriol. 1991 Dec;173(23):7491–7500. doi: 10.1128/jb.173.23.7491-7500.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holck A., Axelsson L., Birkeland S. E., Aukrust T., Blom H. Purification and amino acid sequence of sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Gen Microbiol. 1992 Dec;138(12):2715–2720. doi: 10.1099/00221287-138-12-2715. [DOI] [PubMed] [Google Scholar]
- Holo H., Nilssen O., Nes I. F. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J Bacteriol. 1991 Jun;173(12):3879–3887. doi: 10.1128/jb.173.12.3879-3887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger M. C., Klaenhammer T. R. Cloning, expression, and nucleotide sequence of the Lactobacillus helveticus 481 gene encoding the bacteriocin helveticin J. J Bacteriol. 1990 Nov;172(11):6339–6347. doi: 10.1128/jb.172.11.6339-6347.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993 Sep;12(1-3):39–85. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Koivula T., Sibakov M., Palva I. Isolation and characterization of Lactococcus lactis subsp. lactis promoters. Appl Environ Microbiol. 1991 Feb;57(2):333–340. doi: 10.1128/aem.57.2.333-340.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Moreno F. Genetics of ribosomally synthesized peptide antibiotics. Annu Rev Microbiol. 1992;46:141–163. doi: 10.1146/annurev.mi.46.100192.001041. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lewus C. B., Kaiser A., Montville T. J. Inhibition of food-borne bacterial pathogens by bacteriocins from lactic acid bacteria isolated from meat. Appl Environ Microbiol. 1991 Jun;57(6):1683–1688. doi: 10.1128/aem.57.6.1683-1688.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Hansen J. N. Some chemical and physical properties of nisin, a small-protein antibiotic produced by Lactococcus lactis. Appl Environ Microbiol. 1990 Aug;56(8):2551–2558. doi: 10.1128/aem.56.8.2551-2558.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marugg J. D., Gonzalez C. F., Kunka B. S., Ledeboer A. M., Pucci M. J., Toonen M. Y., Walker S. A., Zoetmulder L. C., Vandenbergh P. A. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, and bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992 Aug;58(8):2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Muriana P. M., Klaenhammer T. R. Cloning, phenotypic expression, and DNA sequence of the gene for lactacin F, an antimicrobial peptide produced by Lactobacillus spp. J Bacteriol. 1991 Mar;173(5):1779–1788. doi: 10.1128/jb.173.5.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriana P. M., Klaenhammer T. R. Conjugal Transfer of Plasmid-Encoded Determinants for Bacteriocin Production and Immunity in Lactobacillus acidophilus 88. Appl Environ Microbiol. 1987 Mar;53(3):553–560. doi: 10.1128/aem.53.3.553-560.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mørtvedt C. I., Nissen-Meyer J., Sletten K., Nes I. F. Purification and amino acid sequence of lactocin S, a bacteriocin produced by Lactobacillus sake L45. Appl Environ Microbiol. 1991 Jun;57(6):1829–1834. doi: 10.1128/aem.57.6.1829-1834.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen-Meyer J., Holo H., Håvarstein L. S., Sletten K., Nes I. F. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992 Sep;174(17):5686–5692. doi: 10.1128/jb.174.17.5686-5692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya R. R., Fremaux C., De Antoni G. L., Klaenhammer T. R. Site-specific integration of the temperate bacteriophage phi adh into the Lactobacillus gasseri chromosome and molecular characterization of the phage (attP) and bacterial (attB) attachment sites. J Bacteriol. 1992 Sep;174(17):5584–5592. doi: 10.1128/jb.174.17.5584-5592.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard G. W., Petzel J. P., van Belkum M. J., Kok J., McKay L. L. Molecular analyses of the lactococcin A gene cluster from Lactococcus lactis subsp. lactis biovar diacetylactis WM4. Appl Environ Microbiol. 1992 Jun;58(6):1952–1961. doi: 10.1128/aem.58.6.1952-1961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagg J. R., Dajani A. S., Wannamaker L. W. Bacteriocins of gram-positive bacteria. Bacteriol Rev. 1976 Sep;40(3):722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema K., Abee T., Haandrikman A. J., Leenhouts K. J., Kok J., Konings W. N., Venema G. Mode of Action of Lactococcin B, a Thiol-Activated Bacteriocin from Lactococcus lactis. Appl Environ Microbiol. 1993 Apr;59(4):1041–1048. doi: 10.1128/aem.59.4.1041-1048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Belkum M. J., Hayema B. J., Jeeninga R. E., Kok J., Venema G. Organization and nucleotide sequences of two lactococcal bacteriocin operons. Appl Environ Microbiol. 1991 Feb;57(2):492–498. doi: 10.1128/aem.57.2.492-498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum M. J., Kok J., Venema G. Cloning, sequencing, and expression in Escherichia coli of lcnB, a third bacteriocin determinant from the lactococcal bacteriocin plasmid p9B4-6. Appl Environ Microbiol. 1992 Feb;58(2):572–577. doi: 10.1128/aem.58.2.572-577.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen J. M., van der Lelie D., Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987 Oct;53(10):2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]