Abstract
Pax genes encode a family of transcription factors, many of which play key roles in animal embryonic development but whose evolutionary relationships and ancestral functions are unclear. To address these issues, we are characterizing the Pax gene complement of the coral Acropora millepora, an anthozoan cnidarian. As the simplest animals at the tissue level of organization, cnidarians occupy a key position in animal evolution, and the Anthozoa are the basal class within this diverse phylum. We have identified four Pax genes in Acropora: two (Pax-Aam and Pax-Bam) are orthologs of genes identified in other cnidarians; the others (Pax-Cam and Pax-Dam) are unique to Acropora. Pax-Aam may be orthologous with Drosophila Pox neuro, and Pax-Bam clearly belongs to the Pax-2/5/8 class. The Pax-Bam Paired domain binds specifically and preferentially to Pax-2/5/8 binding sites. The recently identified Acropora gene Pax-Dam belongs to the Pax-3/7 class. Clearly, substantial diversification of the Pax family occurred before the Cnidaria/higher Metazoa split. The fourth Acropora Pax gene, Pax-Cam, may correspond to the ancestral vertebrate Pax gene and most closely resembles Pax-6. The expression pattern of Pax-Cam, in putative neurons, is consistent with an ancestral role of the Pax family in neural differentiation and patterning. We have determined the genomic structure of each Acropora Pax gene and show that some splice sites are shared both between the coral genes and between these and Pax genes in triploblastic metazoans. Together, these data support the monophyly of the Pax family and indicate ancient origins of several introns.
The Pax genes encode a complex family of transcription factors with multiple DNA-binding domains and diverse functions, and for these reasons, many aspects of their evolution remain speculative. Pax genes are defined by the presence of the paired box, first identified in the Drosophila pair-rule gene paired (1), and encode a large (128-aa) DNA-binding domain (the Paired domain or PD). In addition, a number of Pax genes also encode a complete or partial homeodomain (HD). Pax HDs are distinguished by the presence of a serine residue at position 50 but are clearly related to those encoded by genes such as the Arx and Otx families (2). Pax and related HDs preferentially bind as dimers at palindromic targets consisting of two TAAT half sites (3). The PD consists of two distinct helix–turn–helix motifs (4). In the case of Paired (prd), only the N-terminal PAI subdomain makes DNA contacts (5), but in some other Pax proteins, the C-terminal RED subdomain makes contacts and modulates binding (6, 7). Additional complications in understanding the binding of these proteins are that a distinct set of targets seems to exist for the RED subdomain (8), that complex sites bound by both PD and HDs have been identified (4), and that Pax HDs can heterodimerize with a range of related proteins (3). Nine Pax genes are known in mammals, and eight (nine if eyegone is included) are known in Drosophila; alternative splicing and multiple roles during development complicate the identification of ancestral functions.
Many Pax proteins contain motifs in addition to the PD; most of the arthropod and chordate Pax genes fall into four classes on the basis of comparisons of domain structure and sequences (9–12). The Pax-6 class, which includes Drosophila eyeless, is the only unequivocal case of conservation of function (reviewed in ref. 13). The Pax-2/5/8 class is viewed as that most closely related to the Pax-6 group—the “supergroup” comprising Pax-6/2/5/8 is clearly distinct from the other supergroup, which comprises the Pax-3/7 and Pax-1/9 clades (11). In addition to these four classes that include orthologs from a wide range of animals, many “orphan” Pax genes are known (see, for example, ref. 14), with more restricted distributions.
The approach that we are taking to understanding the evolution of these genes is to characterize the Pax gene complement of the staghorn coral, Acropora millepora, an anthozoan. The Anthozoa are the basal class within the Cnidaria (15–17)—the simplest animals at the tissue level of organization—and are thus likely to reflect ancestral character states more closely than members of the other classes. The rationale behind this assumption is that, in a representative basal animal, genes should be performing more restricted functions, and hence, ancestral roles may be more clearly seen. We previously identified two Pax genes in Acropora (18), one of which has orthologs in the hydrozoans Hydra littoralis (19) and Hydra magnipapillata (20) as well as the scyphozoan Chrysaora quinquecirrha (19). A third cnidarian Pax gene is known from the hydrozoan Podocoryne carnea; EMBL accession no. AJ249563) as well as C. quinquecirrha (19), H. littoralis, and H. magnipapillata (19, 20). Although these genes have provided novel perspectives on Pax gene evolution (18–20), some speculation is involved in relating the known cnidarian genes to the Pax gene classes identified in higher animals. Additionally, nothing is known about the roles of these genes in lower Metazoa.
Herein, we describe two genes from Acropora, bringing the number of Pax genes identified in this animal to four. One of these genes is likely to be orthologous to the Pax-B gene known from several other cnidarians and a single sponge species, and it can be viewed as corresponding to an ancestral Pax-2/5/8 gene; the second falls unambiguously into the Pax-3/7 class and has no orthologs among lower animals. The identification of a Pax-3/7 gene in Acropora indicates that substantial diversification of the Pax family predates the Cnidaria/higher Metazoa split, and the presence of common intron positions is consistent with the monophyly of the Pax family. We have elsewhere argued that Pax diversification accompanied and facilitated the elaboration of the nervous system (2, 18, 21). Consistent with this argument, we show that Pax-Cam is expressed in presumed neurons.
Materials and Methods
Isolation of cDNA Clones.
The construction of the cDNA library is described elsewhere (22). Details of the PCR primers, amplification conditions, and methods used for screening cDNA libraries have been described (18).
Genomic DNA and Genomic Clones.
High molecular weight genomic DNA was isolated from A. millepora egg-sperm bundles as described (23). Size-fractionated genomic DNA that had previously been partially digested with MboI was used for the construction of a genomic library in λGEM12 (Promega) by using the manufacturer's recommended methods. Plaques were screened at moderate stringency by using the homologous cDNA clones as probes. Genomic clones were subjected to direct DNA sequencing with the Applied Biosystems BigDye terminator chemistry, and sequences were determined on ABI310 Genetic Analyzers.
Electrophoretic Mobility-Shift Assay.
A PCR product containing a BamHI site upstream of the complete Pax-Bam paired box followed by a KpnI site and a (TAG) stop codon was generated from the cDNA. This product was cloned into the BamHI and KpnI sites of pQE-30 (Qiagen, Chatsworth, CA), and the recombinant Pax-Bam PD was expressed and purified to homogeneity on Ni+-NTA columns (Qiagen) by using the manufacturer's recommended protocols. Complementary single-stranded oligonucleotides corresponding to the sequence TGGTCACGCTTGAACTATC containing a consensus Pax-2/5/8-binding site were synthesized and annealed by boiling for 5 min followed by cooling to room temperature. Both strands carried 5′ extensions (TT) to enable the incorporation of [α-32P]dATP by Klenow fragment-mediated end filling. Labeled probes were gel purified (JETSorb; GenoMed, GmbH, Bad Oeynhausen, Germany) before use. Binding reactions were carried out by incubation of known amounts of recombinant PD for 30 min at room temperature with 3.5 × 10−10 M double-stranded probe in 20 μl of binding buffer [15 mM Tris/75 mM KCl/0.75 mM EDTA/0.5 mM DTT/0.5 mg/ml BSA/0.05% NP-40/7.5% (vol/vol) glycerol pH 7.5] containing 50 ng/μl poly(dI.dC). Protein–DNA complexes were then analyzed on 6% nondenaturing polyacrylamide gels containing 0.5 × TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3).
Whole-Mount in Situ Hybridization.
A. millepora embryos were fixed at intervals spaced appropriately such that all major morphological stages were represented. Fixation was for 10–20 min in 3.7% (vol/vol) formaldehyde in Millipore-filtered seawater buffered to pH 8.0 with Hepes buffer. Embryos were then washed repeatedly in Millipore-filtered seawater, dehydrated through a graded methanol series, and then stored in absolute methanol at −20°C until needed. The embryos contain large amounts of lipid; thus, a complex delipification procedure must be carried out, which will be described elsewhere (E.E.B., unpublished work). The hybridization solution, hybridization procedure, and in situ probe production have been described by Kucharski et al. (24), except that hybridization was at 55°C. Specimens were cleared through a graded glycerol series and mounted in 90% (vol/vol) glycerol. Photographs were taken on a Zeiss Axioskop with Kodak Ektachrome 64 tungsten film with the resulting images converted to digital form by scanning. Other images were captured directly with a Spot digital camera. Digitized images were processed with adobe photoshop.
Results
Cloning of Acropora Pax Genes.
To search for Acropora Pax genes, PCR was conducted with degenerate primers corresponding to conserved parts of the PD (18). This process led to the identification of four distinct PCR products, each encoding clearly different Pax gene fragments. cDNAs corresponding to each of these genes were isolated from a late embryonic stage cDNA library. Two of these cDNAs have been described (18): one corresponds to Pax-A, previously isolated from both Hydra and the jellyfish Chrysaora (19); the other (Pax-Cam) does not correspond to known cnidarian Pax genes. Further screening led to the identification of two further Acropora Pax genes (see Fig. 1).
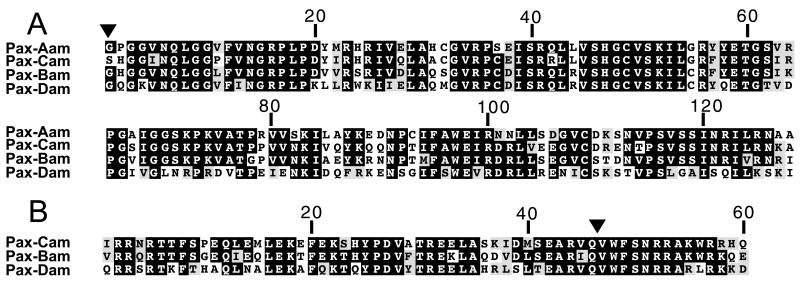
Figure 1.
Alignments of Acropora PD (A) and HD (B) sequences. Shading (generated with boxshade 3.21) indicates identity of at least two (HD) or three (PD) sequences. The inverted triangles indicate two intron positions: that in the first codon of the PD is present in Pax-Aam, Pax-Bam, and Pax-Dam and that in the HD is present in Pax-Bam, Pax-Cam, and Pax-Dam.
Acropora Pax-Bam is likely to be orthologous with Pax-B previously identified in several other cnidarians, whereas Pax-Dam differs substantially from all previously reported cnidarian Pax genes and is most closely related to the Pax-3/7 class (see below). Thus, of the four Acropora Pax genes, two (Pax-Aam and Pax-Bam) have probable orthologs in other cnidarian classes, and two (Pax-Cam and Pax-Dam) either are absent or have not yet been found except in Acropora.
Phylogenetic Relationships of Acropora Pax Proteins.
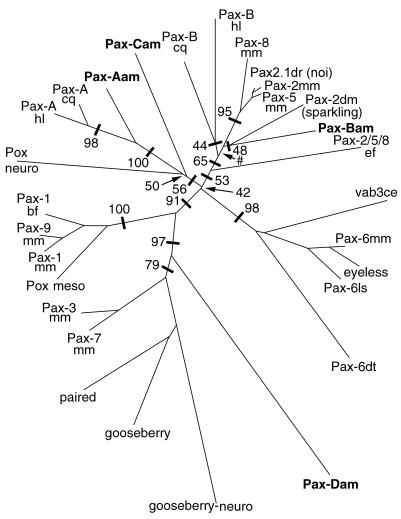
Three of the Acropora Pax genes (Pax-Bam, Pax-Cam, and Pax-Dam) encode complete HDs, and one of these (Pax-Bam) encodes an unambiguous octapeptide motif (see below). The Pax-Aam protein resembles Drosophila Pox neuro (Pox-n) in lacking any trace of a HD and does not seem to contain an octapeptide. To investigate relationships between Pax genes in Acropora, those identified in other cnidarians, and the established Pax classes, phylogenetic analyses were conducted on both the PD and HD sequences. Fig. 2 summarizes results of a number of analyses of PD data.
Figure 2.
Phylogenetic analysis of PD sequences. The tree is shown as an unrooted phylogram and is the result of distance analysis (neighbor-joining method) with paup4b2 (25); 127 rather than 128 amino acids have been used in the analysis, because that is all of the sequence available for Pax-Bcq. Numbers along branches indicate the percentage of 1,000 bootstrap replicates supporting the topology shown. For clarity, some bootstrap values are omitted. The symbol # indicates <40% bootstrap support. The analysis shown includes the sponge sequence sPax-2/5/8 (as Pax-2/5/8ef); note that when this divergent sequence was excluded, bootstrap support for the large clade comprising the cnidarian Pax-B and eumetazoan Pax-2/5/8 sequences increased significantly to 87%. For consistency and simplicity in labeling, Drosophila proteins have retained their original names, but other protein names containing a “Pax-X” in their name have been relabeled according to the formula (Pax) + (designation) + (abbreviation of genus and species). Thus, sPax-2/5/8 from the sponge Ephydatia fluviatilis (20) has been designated Pax-2/5/8ef in the figure.
The cnidarian Pax-A sequences form a distinct and strongly supported clade irrespective of method of analysis. Although bootstrap values within this extended group were somewhat lower, the nearest neighbors of the Pax-A clade are Pax-Cam and Drosophila Pox-n. In our analyses, Pax-Bam fell into a clade containing Hydra (Pax-Bhl) and jellyfish Pax-B (Pax-Bcq), the vertebrate Pax-2/5/8 sequences, and Drosophila sparkling; when the sponge sequence (Pax-Bef) was included in analyses, it was basal to this former clade (Fig. 2). Although some relationships within this large clade are unclear at this stage, our analyses support the assignment of Pax-Bam as orthologous with the synonymous Hydra and jellyfish sequences (see Fig. 2). For example, the PD encoded by Pax-Bam has 101 of 128 identical residues (79% identity) with the H. littoralis sequence (19), and both genes encode unambiguous octapeptides most like those in Pax-2/5/8 proteins (YSINGILG in Acropora; YSISGILG in Hydra). The level of identity between the corresponding HDs (35 of 60), although rather low, should be viewed in the context that the cnidarians are a diverse phylum and that there is possible degeneracy of the Hydra HD (see below). For comparison, the hydrozoan Podocoryne has 112 of 128 and 46 of 60 identities with Hydra in the PD and HD, respectively. In the case of the scyphozoan Chrysaora, no HD data are available, and the identity in the PD is 103 of 127 with Hydra Pax-B—lower identity than between Pax-Bam and Chrysaora Pax-B (106 of 127). Our interpretation of these data is that Pax-B has been subject to fewer constraints than Pax-A and has undergone substantial divergence within the Cnidaria.
The PD encoded by Pax-Dam is clearly related most closely to those of the Pax-3/7 class; in our analyses, the position of Pax-Dam as basal to the Pax-3/7 clade within the Pax-3/7/1/9 supergroup was always strongly supported. In separate analyses (not shown), the Pax-Dam HD fell into the same position (basal to the Pax-3/7 clade); hence, there is consistent support for the view that Pax-Dam corresponds to an ancestral Pax-3/7 gene.
Some Splice Sites Are Conserved Between Acropora and Higher Animals.
To investigate the intron/exon structure of Acropora Pax genes, we isolated genomic clones corresponding to each of the four Pax cDNAs and sequenced these. Perhaps not surprisingly, the Acropora genomic loci were, in general, significantly less complex than those of many of the vertebrate or Drosophila Pax genes. The Pax-Dam locus seems to be the least complex, consisting of four exons extending over a little under 4 kilobases; Pax-Aam and Pax-Cam each consist of five exons over approximately 8 kilobases. The Pax-Bam locus was more complex, comprising nine exons spread over approximately 12 kilobases (I.S., unpublished work). Comparison of the intron/exon organization led to the identification of two intron positions common among the Acropora Pax genes and between these and many other Pax genes. We have previously reported that Pax-Cam contains an intron at a position corresponding to residues 46/47 in the HD (18); an intron is also present at this position in both Pax-Bam and Pax-Dam. Additionally, an intron is present in the first codon of the paired box in Pax-Aam, Pax-Bam, Pax-Dam (see Fig. 1), and a range of other Pax genes. Pax-Cam has an intron at a similar, but not identical, position—in this case, 5′ of the paired box. No data are available for the hydra genomic loci. Sun et al. (19) report the apparent absence of introns in jellyfish Pax-A and Pax-B paired boxes.
The Pax-Bam PD Binds to Consensus Pax-2/5/8-Binding Sites.
Like members of the Pax-2/5/8 class, but unlike the Pax-A/Pox-n and Pax-6 classes, cnidarian Pax-B PDs each have Q-R--H at positions 42, 44, and 47, which are known to be critical in determining DNA-binding specificity (26). This arrangement implies that Pax-B should bind Pax-2/5/8 target sites. The DNA-binding ability of the Pax-Bam PD was evaluated in electrophoretic mobility-shift assays by using oligonucleotides containing the consensus Pax-2/5/8-binding site TGGTCACGCTTGA (26, 27). Recombinant Pax-Bam PD bound to the consensus binding site with high affinity (see Fig. 3); under the conditions used (3.5 × 10−10 M probe and 10,000-fold excess of competitor DNA), >87% of probe bound with 10−7 M protein and the apparent Kd was calculated as 5 × 10−9 M.
Figure 3.
DNA-binding assay. (A) The binding of recombinant Pax-Bam PD to oligonucleotides containing a consensus Pax-2/5/8-binding site was determined by electrophoretic mobility-shift assays. S and F indicate the positions of shifted and free probe, respectively. Concentration of oligonucleotide was held constant (3.5 × 10−10 M), and the concentration of PD varied from 10−6 to 10−9 M; the panel on the extreme right represents negative control (no added protein). (B) Quantitation of DNA-binding. The histogram shows the percentage of probe shifted in relation to the amount of PD added. Raw data from A were quantified on a Molecular Dynamics Storm PhosphorImager by using the imagequant nt software.
Pax-Cam Is Expressed in Presumed Neurons During Development.
Although Pax genes have many specialized and diverse roles in higher animals, most are expressed in the nervous system during development. The location of this expression implies that the ancestral role of Pax genes (at least those encoding HDs) was in the nervous system. To test this hypothesis, we examined the distribution of Pax-Cam mRNA during Acropora embryogenesis by using whole-mount in situ hybridization (Fig. 4). Pax-Cam was selected for this purpose, because, of the cnidarian genes, it most closely reflects the ancestor of the Pax-6 genes (see below and ref. 18).
Figure 4.
Localization of Pax-Cam mRNA by in situ hybridization. (A) Photomontage showing Pax-Cam expression in scattered transectodermal cells in a pear-shaped planula larva. These cells are more abundant at the aboral (a) than the oral (o) end. Arrows identify some of these strongly expressing ectodermal cells. The line of white dots marks the basement membrane separating ectoderm from gastroderm (endoderm). Staining of occasional cells in the gastroderm is believed to be nonspecific. (B) Two cells with nuclei (n) midway across the ectoderm appear to project to a clump of expressing cells on the basement membrane. The message is excluded from the nuclei. (C) A single labeled cell with a basal nucleus (n) projects across the ectoderm. (Bars = 100 μm for A and 10 μm for B and C.)
The Pax-Cam message is detectable at 36 h (late in gastrulation) and is expressed more abundantly at 48 h (J.C., unpublished work), by which time the embryo is pear-shaped (see figure 4 in ref. 28). Pax-Cam in situ preparations show scattered labeled transectodermal cells (Fig. 4A). The morphology of these cells is both consistent with their assignment as neurons and inconsistent with other cell types described from the anthozoan ectoderm (29). However, unequivocal identification is not yet possible because panneural markers, recognized by either antibody or in situ hybridization probes, have not yet been developed for Acropora or most other cnidarians. Some of these presumed neurons have their nuclei midway across the ectoderm (Fig. 4B), whereas others have basal nuclei (Fig. 4C). Our in situ preparations also show occasional clumps of stained cells near the basement membrane. Fig. 4B shows expressing cells with nuclei halfway across the ectoderm apparently projecting to one of these clumps.
Discussion
The identification of four distinct Pax genes in a basal cnidarian and the apparent relatedness of some of these to the classes known from higher animals significantly refine our ideas about the evolution of this complex family of genes. Whereas a degree of uncertainty is involved in relating other cnidarian genes to the established Pax classes, the identification of an unambiguous Pax-3/7 ortholog in Acropora (Pax-Dam) clearly indicates that substantial diversification of Pax genes had already occurred before the Cnidaria/higher Metazoa split.
The Pax sequences reported herein and the structural data for the Acropora genomic loci are consistent with the monophyly of the Pax family. The possession of common splice sites has frequently been used to support common ancestry, and two intron positions are common in the Acropora Pax genes—that at positions 46/47 in the HD and in the first codon of the PD. The first of these is shared by a wide range of Pax and paired-like genes (18). The PD N-terminal intron present in three of the four coral genes is also present in a diverse range of vertebrate and invertebrate Pax genes, including many Pax-6 (13) and Pax-2/5/8 (30) genes. These introns are clearly very old, predating the cnidarian/triploblast split (at least 543 million years ago; ref. 31), and can be considered to provide further support for the notion of monophyly of the Pax gene family.
In terms of affinity with the known classes, Pax-Dam is the least ambiguous of all of the cnidarian Pax genes identified to date—both its PD and HD clearly assign it to the Pax-3/7 class. Unlike some of its orthologs, Pax-Dam does not encode an octapeptide; however, this motif is not diagnostic of the Pax-3/7 class (2). Like other Pax-3/7 class genes, the HD encoded by Pax-Dam has a serine residue at position 4. The identification of a Pax-3/7 class gene in a basal metazoan has implications for the evolution of function in this group. By analogy, it is likely that Pax-Dam is involved in cell-fate specification in the nervous system, because common patterns of expression in vertebrates, Drosophila, and an ascidian imply this kind of original role for Pax-3/7 genes (32). The absence of any indication of repetitive or segmental organization in cnidarians implies that such a mode of Pax-3/7 expression, seen in Drosophila (prd and gsb) and in the ascidian (33), is derived rather than ancestral.
Our analyses of PD data support the view that cnidarian Pax-B is a “primitive” representative of the Pax-2/5/8 class. Whereas the cnidarian Pax-B genes are typical of this class in encoding octapeptides, they are atypical in that they encode complete (rather than incomplete) HDs. However, we note that both of the Hydra Pax-B proteins are predicted to contain proline residues (Pro-43) in helix 3 of the HD (Ala in almost all other HD proteins), a substitution likely to prevent dimerization and perhaps also binding at P2/P3 sites. Effectively, Hydra Pax-B can be thought of as on its way to becoming a true Pax-2/5/8 gene, because the HD that it encodes may be unable to bind DNA and is therefore likely to be lost. Unlike that in Hydra, the Acropora Pax-Bam HD has an alanine residue at (HD) position 43, and should therefore behave as a typical Pax HD. DNA-binding experiments indicate that the Pax-Bam HD binds P2 and P3 sites (I.S., unpublished work), but the ability of the protein to dimerize has not yet been clarified.
Although our recently acquired Acropora data are consistent with our previous model for the evolution of the Pax classes (18), we have had to reexamine it in the light of the publication of a sponge Pax sequence (sPax-2/5/8 = Pax-2/5/8ef in Fig. 2; ref. 20). We previously put forward the idea that the acquisition of a homeobox was a key event in Pax gene evolution, allowing a transition from general roles in cell-fate specification to more specific functions in anterior patterning (18). We also raised the possibility of sponges containing Pax genes but, because of an implied link with nervous system patterning, considered it unlikely that these would encode HDs. In apparent contradiction to this hypothesis, a gene most closely related to the Pax-2/5/8 class was recently identified in the freshwater sponge E. fluviatilis (20). Surprisingly, the E. fluviatilis gene encodes a complete although substantially degenerate HD.
Previously, we suggested that Pax-A and Pax-C represent the “before” and “after” states with respect to the gene fusion event. Consistent with this suggestion, Pax-Aam and Pax-Cam remain the most closely related of the four Acropora Pax proteins in their PD sequences. The high degree of identity between Pax-Bam and Pax-Cam in both PD and HD sequences implies a common origin via duplication, and Pax-Bam seems to be an ancestral Pax-2/5/8 gene. These implied relationships lead to the scheme shown as Fig. 5A, which is modified from that of Catmull et al. (18).
Figure 5.
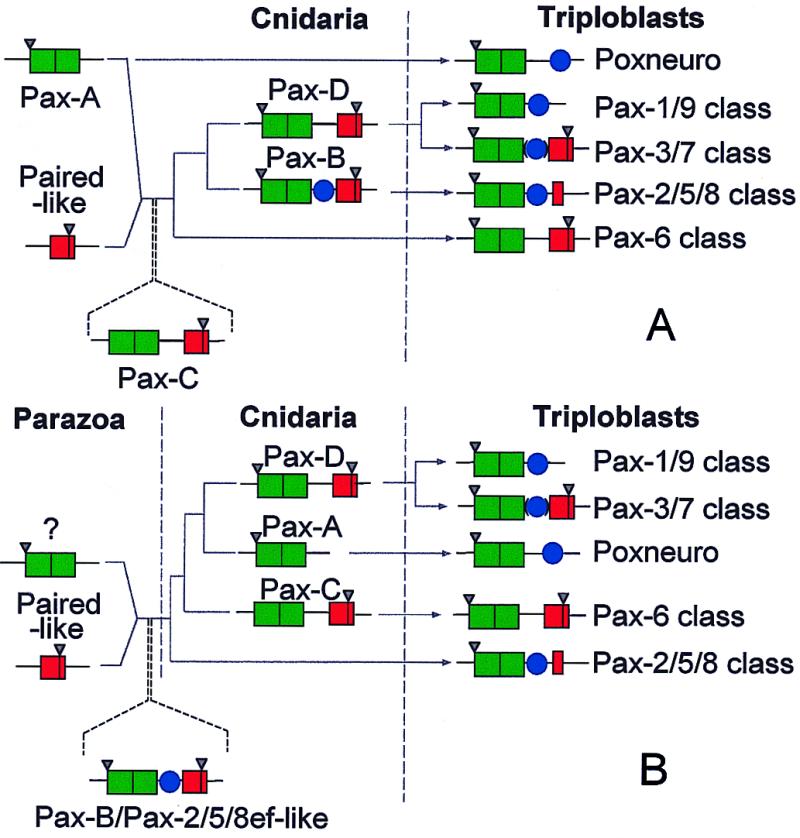
Alternative models for evolution of the Pax classes. (A) This model is based on our previous scheme (18) but accommodates the data reported in this study. Herein, we view Pax-Aam and Pax-Cam as representing the before and after states of a fusion event between a Pax-A/Pox-n-like gene and a paired-like homeobox gene. (B) An alternative model, in which Pax-B/sPax-2/5/8 (rather than Pax-Cam) is seen as reflecting the ancestor of most of the metazoan Pax genes. The models also differ in interpreting the Pax-A/Pox-n class as ancestral (A) or derived (B). In both schemes, Pax-Cam corresponds to the ancestor of the Pax-6 class, and Pax-Dam the precursor of the Pax-3/7 class; the Pax-1/9 class is related to the Pax-3/7 class via loss of the homeobox. Common ancestry of Pax genes is implied by the fact that all of the cnidarian Pax HDs are intermediate between those of the Pax-6 and Pax-3/7 classes (although that in Pax-Dam is very close to the latter class) and share splice sites. Pax-Aam, Pax-Bam, and Pax-Dam have an intron at positions corresponding to the first codon of the PD that is also present in a number of other Pax genes, and each of the Acropora Pax homeoboxes contains an intron at a position corresponding to amino acids 46/47 of the HD (these are indicated by inverted triangles in the scheme). Green box, PD; red box, HD; blue circle, octapeptide.
If we accept the idea of a single origin of Pax genes, then the gene fusion event must have predated the Porifera/Metazoa split, by which time the precursor of the Pax-2/5/8 class was distinct. Although the sponge data can be accommodated by our previous model of Pax evolution, such an accommodation requires that there have been either substantial gene losses or the existence of undetected gene classes in the Porifera. An alternative evolutionary scheme, which is also compatible with all of the available data, is shown in Fig. 5B. Under this scheme, Pax-B (rather than Pax-C in our previous model) is seen as reflecting the ancestor of most of the metazoan Pax genes; although Pax-C is derived, it most closely reflects the ancestor of the Pax-6 genes. A key difference between the two models is that in the latter (Fig. 5B), Pax-A is seen as derived via loss of the HD, whereas in the former (Fig. 5A), it is viewed as ancestral (i.e., it contained only a PD and had never gained a HD). In the absence of an appropriate outgroup, it is not possible to decide which direction evolution has taken—which of the Pax types from lower animals most resembles the ancestor of the extant genes. This issue can be resolved only by more comprehensive sampling of lower animals; expression patterns of the genes are likely to be highly informative with respect to ancestral roles and conserved functions.
Acknowledgments
D.J.M. and J.C. were supported by grants from the Australian Research Council. J.R.-H. was supported by an Australian Postgraduate Research Award and an Australian National University Collaborative Scholarship. P.C. was supported by the Collen Foundation (Belgium) and the Sandoz Foundation (Switzerland).
Abbreviations
- PD
Paired domain
- HD
homeodomain
Footnotes
References
- 1.Frigerio G, Burri M, Bopp D, Baumgartner S, Noll M. Cell. 1986;47:735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- 2.Galliot B, de Vargas C, Miller D J. Dev Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- 3.Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 4.Jun S, Desplan C. Development (Cambridge, UK) 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 5.Xu W, Rould M A, Jun S, Desplan C, Pabo C O. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- 6.Czerny T, Schaffner G, Busslinger M. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 7.Xu H E, Rould M A, Xu W, Epstein J A, Maas R L, Pabo C O. Genes Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein J A, Glaser T, Cai J, Jepeal L, Walton D S, Maas R L. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 9.Noll M. Curr Opin Genet Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- 10.Bürglin T R. In: Guidebook to the Homeobox Genes. Duboule D, editor. Oxford: Oxford Univ. Press; 1994. pp. 27–71. [Google Scholar]
- 11.Balczarek K A, Lai Z C, Kumar S. Mol Biol Evol. 1997;14:829–842. doi: 10.1093/oxfordjournals.molbev.a025824. [DOI] [PubMed] [Google Scholar]
- 12.Miller D J. Zool Stud. 1999;38:367–372. [Google Scholar]
- 13.Callaerts P, Halder G, Gehring W J. Annu Rev Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- 14.Hobert O, Ruvkun G. Trends Genet. 1999;15:214–216. doi: 10.1016/s0168-9525(99)01731-x. [DOI] [PubMed] [Google Scholar]
- 15.Bridge D, Cunningham C W, Schierwater B, Desalle R, Buss L W. Proc Natl Acad Sci USA. 1992;89:8750–8753. doi: 10.1073/pnas.89.18.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bridge D, Cunningham C W, DeSalle R, Buss L W. Mol Biol Evol. 1995;12:679–689. doi: 10.1093/oxfordjournals.molbev.a040246. [DOI] [PubMed] [Google Scholar]
- 17.Odorico D M, Miller D J. Proc R Soc London Ser B. 1997;264:77–82. doi: 10.1098/rspb.1997.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catmull J, Hayward D C, Macintyre N E, Reece-Hoyes J S, Mastro R, Callaerts P, Ball E E, Miller D J. Dev Genes Evol. 1998;208:352–356. doi: 10.1007/s004270050191. [DOI] [PubMed] [Google Scholar]
- 19.Sun H, Rodin A, Zhou Y, Dickinson D P, Harper D E, Hewett-Emmett D, Li W H. Proc Natl Acad Sci USA. 1997;94:5156–5161. doi: 10.1073/pnas.94.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoshiyama D, Suga H, Iwabe N, Koyanagi M, Nikoh N, Kuma K, Matsuda F, Honjo T, Miyata T. J Mol Evol. 1998;47:640–648. doi: 10.1007/pl00006421. [DOI] [PubMed] [Google Scholar]
- 21.Galliot B, Miller D J. Trends Genet. 2000;16:1–5. doi: 10.1016/s0168-9525(99)01888-0. [DOI] [PubMed] [Google Scholar]
- 22.Brower D L, Brower S M, Hayward D C, Ball E E. Proc Natl Acad Sci USA. 1997;94:9182–9187. doi: 10.1073/pnas.94.17.9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMillan J, Yellowlees D, Heyward A, Harrison P, Miller D J. Mar Biol. 1988;98:271–276. [Google Scholar]
- 24.Kucharski R, Ball E E, Hayward D C, Maleszka R. Gene. 2000;242:399–405. doi: 10.1016/s0378-1119(99)00503-x. [DOI] [PubMed] [Google Scholar]
- 25.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 1998. , Version 4. [Google Scholar]
- 26.Czerny T, Busslinger M. Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epstein J A, Cai J, Glaser T, Jepeal L, Maas R L. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 28.Miller D J, Ball E E. BioEssays. 2000;22:291–296. doi: 10.1002/(SICI)1521-1878(200003)22:3<291::AID-BIES11>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 29.Fautin D G, Mariscal R N. In: Microscopic Anatomy of Invertebrates: Vol. 2 Placozoa, Porifera, Cnidaria, and Ctenophora. Harrison F W, Westfall J A, editors. New York: Wiley–Liss; 1991. pp. 267–358. [Google Scholar]
- 30.Pfeffer P L, Gerster T, Lun K, Brand M, Busslinger M. Development (Cambridge, UK) 1998;125:3063–3074. doi: 10.1242/dev.125.16.3063. [DOI] [PubMed] [Google Scholar]
- 31.Grotzinger J P, Bowring S A, Saylor B Z, Kaufman A J. Science. 1995;270:598–604. [Google Scholar]
- 32.Wada H, Saiga H, Satoh N, Holland P W H. Development (Cambridge, UK) 1998;125:1113–1122. doi: 10.1242/dev.125.6.1113. [DOI] [PubMed] [Google Scholar]
- 33.Wada H, Holland P W, Sato S, Yamamoto H, Satoh N. Dev Biol. 1997;187:240–252. doi: 10.1006/dbio.1997.8626. [DOI] [PubMed] [Google Scholar]