Abstract
It has been proposed that food animals represent the source of glycopeptide resistance genes present in enterococci from humans. We demonstrated the transfer of vanA and of other resistance genes from porcine to human Enterococcus faecium at high frequency in the digestive tract of gnotobiotic mice. Tylosin in the drinking water favored colonization by transconjugants.
The acquisition and spread of glycopeptide-resistant enterococci (GRE) is a global problem, although the selective pressure that has led to dissemination differs between geographical areas (19). In the United States, where antibiotics which represent high-level risk factors for colonization or infection by GRE are extensively prescribed, such strains are isolated from hospitalized patients but not from community-based volunteers without hospital exposure or from the environment or animals (4, 10). By contrast, a low incidence of clinical GRE infection (2.2%) is observed in most European countries (8) but the strains are found in the healthy human population and in animals (1). As opposed to practices in the United States, the glycopeptide avoparcin was used as a growth promoter in animal husbandry in Europe until 1997 and is associated with the dissemination (in poultry and pigs in particular) of enterococci that are cross-resistant to avoparcin and vancomycin (9). The finding of GRE in nonhospitalized humans and in meat eaters but not in vegetarians further suggests a food-associated spread of vancomycin-resistant enterococci from animals to humans (17). A study after ingestion of GRE of animal origin by healthy volunteers revealed the presence of such strains in human stools for prolonged periods of time (18). The entry of GRE of animal origin into the human food chain not only allows these strains to become established in the human gut but can also favor transfer of their resistance genes to human commensals (21). It has been shown that in 20 GRE isolates taken in Germany from infections in patients, from nonhospitalized humans, from sewage and animal feces, and from meat products, the vanA operon was structurally conserved, which suggests gene spread among these various ecosystems (22). Transfer of genes encoding glycopeptide resistance between animal and human strains in nature is still controversial (15), even though transfer of the satA gene (encoding resistance to streptogramin A) has been demonstrated for Enterococcus faecium in the gastrointestinal tract of gnotobiotic rats (11). The purpose of this study was to examine the possibility of the transfer of antibiotic resistance genes between E. faecium strains of animal and human origins in vitro and in vivo in the digestive tract of gnotobiotic mice.
Human fecal isolate E. faecium 64/3 (resistant to rifampin and fusidic acid) was used as a recipient. Four E. faecium isolates of porcine origin (UW4, UW7, UW261, and UW262), harboring the vanA and ermB genes (conferring resistance to vancomycin and erythromycin, respectively), the tet(L) and ant(6) genes (mediating resistance to tetracycline and streptomycin, respectively, in UW262), and the tet(M) tetracycline determinant (in UW7), were used as donors. Transposon Tn1546, which carries the vanA gene cluster in all animal strains, was characterized by PCR as previously described (12).
In vitro transfer.
Conjugal transfer of resistance was tested by filter mating on agar (5). Selective medium, containing vancomycin (50 μg/ml) for isolation of the donor, rifampin (30 μg/ml) and fusidic acid (20 μg/ml) for enumeration of the recipient, and rifampin, fusidic acid, and vancomycin, erythromycin (10 μg/ml), tetracycline (50 μg/ml), or streptomycin (1 mg/ml) for detection of the transconjugants, was used. Whatever the donor strain, transfer of the vanA and ermB genes was obtained (Table 1). Average transfer frequencies in three independent experiments for each donor ranged from 9 × 10−5 to 19 × 10−2. Transconjugants were assigned to one of three groups on the basis of their phenotypes and genotypes. The presence of the resistance genes was studied in a multiplex assay using primers that allow amplification of vanA (6), ermB (2), ant(6) (20), tet(L) (1), and tet(M) (16). Amplification consisted of 30 cycles of denaturation at 94°C for 45 s, annealing at 47°C for 45 s, and elongation at 72°C for 1 min. Transconjugants of type I were resistant to vancomycin and erythromycin following acquisition of the vanA and ermB genes. The type II strains were resistant to vancomycin (vanA) and susceptible to erythromycin, whereas type III transconjugants were resistant to erythromycin (ermB) and susceptible to vancomycin. The transconjugants obtained in vitro were all of type I irrespective of the selective medium. Transconjugants from UW262 obtained in vitro were analyzed for resistance to tetracycline [tet(L)] and streptomycin [ant(6)]. All transconjugants tested were resistant to streptomycin (transfer frequency, 5 × 10−5), and some of them also acquired resistance to tetracycline (transfer frequency, 7 × 10−8). The in vitro transconjugants from UW7 were analyzed for resistance to tetracycline [tet(M)]. Tetracycline resistance was transferred at a frequency of 10−6.
TABLE 1.
In vitro transfer of the vanA gene cluster to E. faecium 64/3
| E. faecium donor | Mean (± SD) transfer frequencya with selection on:
|
|
|---|---|---|
| Vancomycin (50 μg/ml) | Erythromycin (10 μg/ml) | |
| UW261 | 9 (± 0.8) × 10−5 | 1 (± 0.1) × 10−4 |
| UW262 | 8 (± 3.5) × 10−4 | 4 (± 2.8) × 10−3 |
| UW4 | 8 (± 5.9) × 10−2 | 2 (± 6.8) × 10−1 |
| UW7 | 3 (± 2.4) × 10−4 | 8 (± 6.1) × 10−4 |
Number of transconjugants relative to the number of donors after the mating period; results represent averages of a minimum of three experiments.
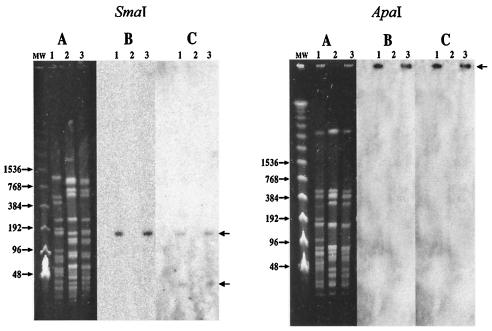
Gene transfer was confirmed by pulsed-field gel electrophoresis (PFGE) (13). The vanA (7) and ermB (3) probes were labeled with [α-32P]dCTP by random priming using a commercially available kit (Amersham Biosciences, Orsay, France). The SmaI- and ApaI-generated patterns of the strain UW262 donor and of the strain 64/3 recipient were markedly different (Fig. 1). The SmaI pattern of a randomly selected transconjugant differed from that of the 64/3 recipient by two additional bands of ca. 140 kb and of less than 48 kb that were present in the donor. The 140-kb band hybridized with the vanA probe, and both bands hybridized with ermB. The ApaI patterns of the recipient and of the transconjugant were indistinguishable, and hybridization of the probes was performed with the DNA remaining in the well. Taken together, these results indicate that a conjugative plasmid of ca. 180 kb with two SmaI but not ApaI cutting sites carries both the vanA and ermB genes.
FIG. 1.
PFGE analysis of SmaI (left panel)- or ApaI (right panel)-digested genomic DNA of E. faecium. (A) Lanes: MW, molecular weight markers; 1, strain UW262; 2, strain 64/3; 3, strain 2T2301VA. The results of Southern hybridization of the gels shown in panels A with 32P-labeled vanA (B) and ermB (C) probes are presented. The sizes of the markers (in kilobase pairs) are indicated on the left.
We observed a three- to fivefold increase in transfer frequency when selection of the transconjugants was on erythromycin instead of on vancomycin (Table 1). This difference may be due to the fact that macrolide resistance is readily expressed phenotypically in the transconjugants, since it is constitutive and due to methylation of a single adenine residue in 23S rRNA. By contrast, VanA-type glycopeptide resistance is slowly inducible and requires remodeling of the entire bacterial cell wall (3). Selection of erythromycin-resistant enterococci may therefore lead to that of GRE, the ermB and vanA genes often being carried by the same transferable genetic element (14). Thus, in addition to cross-resistance, coresistance should be taken into account in determining antibiotic usage policy.
In vivo conjugation.
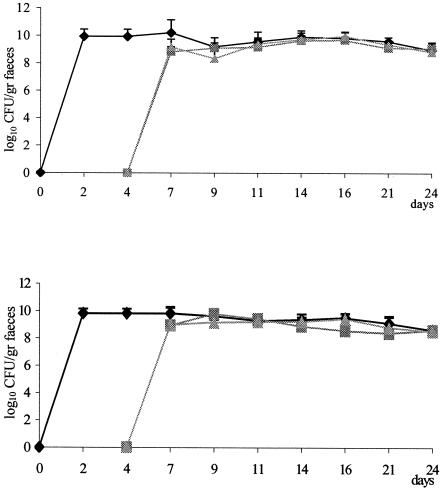
Germfree consanguineous C3H mice supplied by INRA (Jouy-en-Josas, France) and maintained in separate isolators (J.C.E. Biotechnology, Vichy, France) were fed ad libitum with a commercial diet sterilized by gamma irradiation (4 Mrad). Every mouse received 108 CFU of recipient strain 64/3 intragastrically in a volume of 600 μl and, 4 days later (after verification of the implantation of the recipient strain), 108 CFU of the donor in a volume of 600 μl. The strain UW7 donor, which had a frequency of mutation to rifampin and fusidic acid resistance lower than 1 × 10−12 and a high vancomycin resistance transfer frequency of 3.3 × 10−4, was selected for the in vivo experiments. The first experiment studied the evolution of bacterial populations within mouse feces, lasted 24 days, and included five mice in each of two groups: a control group that received drinking water without antibiotic and a treated group that received drinking water supplemented with 0.1 μg of tylosin/ml. Geometric averages of bacterial counts in the two groups are shown in Fig. 2. In the first sample (collected 3 days after donor inoculation), transconjugants were detected at concentrations ranging from 9 to 10 log10 CFU per gram of feces and established at population levels equal to those of the recipient and of the donor throughout the experiment. Thus, there was no apparent ecological disadvantage for the transconjugant population relative to the other populations following the acquisition of exogenous genetic information. This may be explained, at least in part, by the fact that the transconjugants corresponded to already established recipients that had acquired the vanA operon and not to a new colonizing strain. The in vivo transconjugants were of the three types previously presented in vitro, with a majority (86%) belonging to type I and 8% and 6% to types II and III, respectively. Tetracycline resistance [tet(M)] was detected in transconjugants starting at day 8. These data, along with those from PFGE analysis, revealed that the vanA and ermB genes were located on the same large self-transferable plasmid. The tetracycline resistance gene tet(M) may be located in the host chromosome as part of a conjugative transposon.
FIG. 2.
Bacterial counts in homogenized feces of gnotobiotic mice supplied with pure water (top panel) or with water supplemented with 0.1 μg of tylosin/ml (bottom panel). Germfree mice were inoculated on day 0 with 108 E. faecium 64/3 recipients and on day 4 with 108 E. faecium UW7::Tn1546 donors. Symbols: ♦, E. faecium 64/3; ▪, E. faecium UW7::Tn1546; ▴, E. faecium 64/3::Tn1546.
To study the bacterial populations in situ in the digestive tract, all mice were sacrificed at day 24, the intestinal tracts (from pylori to rectum) were removed, weighted, diluted 10-fold, and homogenized with an Ultraturax mixer (Bioblock, Illkirch Cedex, France), and dilutions were plated on brain heart infusion agar supplemented with antibiotics. Enumeration averages of bacterial populations within the digestive tracts after the sacrifice of both groups of mice at day 24 were close to those obtained from fecal samples (Table 2).
TABLE 2.
Bacterial counts in homogenized intestines of gnotobiotic mice
| E. faecium strain or transconjugant | Bacterial count in drinking water with:
|
|
|---|---|---|
| No antibiotic | Tylosin (0.1 μg/ml) | |
| UW7 | 1 × 109 | 3 × 108 |
| 64/3 | 4 × 108 | 1 × 108 |
| VanR transconjugant | 4 × 107 | 9 × 108a |
α ≤ 5% in the Mann-Whitney U test in comparison to transconjugants in mice untreated with tylosin.
The second experiment studied the kinetics of gene transfer 5, 10, and 24 h following donor inoculation. In vivo gene transfer occurred at an unexpectedly high rate, since at 5 h after donor inoculation, transconjugants could already be isolated from all mice with counts of 105 CFU per gram of feces. This differs from the results of transfer of the satA gene within the digestive tract of gnotobiotic rats, during which transconjugants were observed only 3 days after donor inoculation (11). Early transfer of the vanA operon suggests that even a brief transit of enterococci of animal origin would allow resident human bacteria to acquire glycopeptide (as well as other) resistance genes.
We investigated the impact of subinhibitory concentrations of tylosin on the transfer of the vanA operon. Tylosin was not associated with a significant increase of in vitro transfer of vancomycin resistance (data not shown). However, tylosin significantly raised levels of colonization by transconjugants in treated mice (≤5% in a Mann-Whitney Utest) (Table 2). These results are reminiscent of those of an in vivo study in which the use of tetracycline as a selective agent in gnotobiotic mice raised the transfer frequency of Tn1545 from E. faecalis to Listeria monocytogenes by 20-fold (5). Thus, the presence of antibiotic residues in the gastrointestinal tract may favor gene transfer from animal to human strains.
Enterococci that harbor antibiotic resistance genes are common in the digestive tract of animals. It has been suggested that these bacteria might serve as a reservoir of resistance genes for human digestive microflora. Previous investigations have revealed the occurrence of horizontal gene transfer between bacteria that colonize livestock and those that colonize humans. In this study, we have shown that various resistance genes can be conjugatively transferred from an E. faecium strain of animal origin to a human strain of the same species in the gastrointestinal tracts of gnotobiotic mice in the absence of selective pressure. The ease with which we were able to obtain gene transfer from animal to human enterococci suggests that gene exchange under natural conditions might take place more commonly than previously suspected.
Acknowledgments
We thank H. Boureau and G. Gerbaud for technical advice to C.M. and W. Witte for the gift of the strains.
C.M. was supported in part by a Nutrition Research Grant from Comité Interprofessionnel de la Dinde Française.
REFERENCES
- 1.Aarestrup, F. M., Y. Agerso, P. Gerner-Smidt, M. Madsen, and L. B. Jensen. 2000. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 37:127-137. [DOI] [PubMed] [Google Scholar]
- 2.Angot, P., M. Vergnaud, M. Auzou, R. Leclercq, and Observatoire de Normandie du Pneumocoque. 2000. Macrolide resistance phenotypes and genotypes in French clinical isolates of Streptococcus pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 19:755-758. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, M., and P. Courvalin. 1986. Contribution of two different mechanisms to erythromycin resistance in Escherichia coli. Antimicrob. Agents Chemother. 30:694-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coque, T. M., J. F. Tomayko, S. C. Ricke, P. C. Okhyusen, and B. E. Murray. 1996. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob. Agents Chemother. 40:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doucet-Populaire, F., P. Trieu-Cuot, I. Dosbaa, A. Andremont, and P. Courvalin. 1991. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob. Agents Chemother. 35:185-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinical relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutka-Malen, S., C. Molinas, M. Arthur, and P. Courvalin. 1990. The VANA glycopeptide resistance protein is related to D-alanyl:D-alanine ligase cell wall biosynthesis enzymes. Mol. Gen. Genet. 224:364-372. [DOI] [PubMed] [Google Scholar]
- 8.Felmingham, D., D. F. J. Brown, and C. J. Soussy. 1998. The European glycopeptide resistance survey of gram-positive bacteria for 1995. Diagn. Microbiol. Infect. Dis. 31:563-571. [DOI] [PubMed] [Google Scholar]
- 9.Gambarotto, K., M. C. Ploy, F. Dupron, M. Giangiobbe, and F. Denis. 2001. Occurrence of vancomycin-resistant enterococci in pork and poultry products from a cattle-rearing area of France. J. Clin. Microbiol. 39:2354-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbarth, S., S. Cosgrove, and Y. Carmeli. 2002. Effects of antibiotics on nosocomial epidemiology of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 46:1619-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobsen, B. L., M. Skou, A. M. Hammerum, and L. B. Jensen. 1999. Horizontal transfer of the satA gene encoding streptogramin A resistance between isogenic Enterococcus faecium strains in the gastrointestinal tract of gnotobiotic rats. Microb. Ecol. Health Dis. 11:241-247. [Google Scholar]
- 12.Ligozzi, M., C. G. Lo, and R. Fontana. 1998. vanA gene cluster in a vancomycin-resistant clinical isolate of Bacillus circulans. Antimicrob. Agents Chemother. 42:2055-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noble, W. C., Z. Virani, and R. G. A. Cree. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 20:91-98. [DOI] [PubMed] [Google Scholar]
- 15.Phillips, I. 1999. The use of bacitracin as a growth promoter in animals produces no risk to human health. J. Antimicrob. Chemother. 44:725-728. [DOI] [PubMed] [Google Scholar]
- 16.Roberts, M. C., Y. Pang, D. E. Riley, S. L. Hillier, R. C. Berger, and J. N. Krieger. 1993. Detection of Tet M and Tet O tetracycline resistance genes by polymerase chain reaction. Mol. Cell. Probes 7:387-393. [DOI] [PubMed] [Google Scholar]
- 17.Schouten, M. A., A. Voss, and J. A. Hoogkamp-Korstanje. 1997. VRE and meat. Lancet 349:1258. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen, T. L., M. Blom, D. L. Monnet, N. Frimodt-Moller, R. L. Poulsen, and F. Espersen. 2001. Transient intestinal carriage after ingestion of antibiotic-resistant Enterococcus faecium from chicken and pork. N. Engl. J. Med. 345:1161-1166. [DOI] [PubMed] [Google Scholar]
- 19.Sundsfjord, A., G. S. Simonsen, and P. Courvalin. 2001. Human infections caused by glycopeptide-resistant Enterococcus spp.: are they a zoonosis? Clin. Microbiol. Infect. 7(Suppl. 4):16-33. [DOI] [PubMed] [Google Scholar]
- 20.Swenson, J. M., M. J. Ferraro, D. F. Sahm, N. C. Clark, D. H. Culver, F. C. Tenover, and the National Committee for Clinical Laboratory Standards Study Group onEnterococci. 1995. Multilaboratory evaluation of screening methods for detection of high-level aminoglycoside resistance in enterococci. J. Clin. Microbiol. 33:3008-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Bogaard, A. E., R. Willems, N. London, J. Top, and E. E. Stobberingh. 2002. Antibiotic resistance of faecal enterococci in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 49:497-505. [DOI] [PubMed] [Google Scholar]
- 22.Werner, G., I. Klare, and W. Witte. 1997. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol. Lett. 155:55-61. [DOI] [PubMed] [Google Scholar]