Abstract
It is generally thought that there is full cross-resistance in Mycobacterium tuberculosis between the aminoglycoside drugs kanamycin and amikacin. However, kanamycin resistance and amikacin susceptibility were seen in 43 of 79 (54%) multidrug-resistant Estonian isolates, indicating that there might be a need to test the resistance of M. tuberculosis isolates to both drugs.
Since the recognition of the remarkable activity of streptomycin (SM) against Mycobacterium tuberculosis in 1944 (2), aminoglycosides have been a major component of therapy for tuberculosis. Kanamycin (KM) and the closely related amikacin (AK) are commonly used for treatment of multidrug-resistant tuberculosis (MDR-TB) (11). Resistance to SM in M. tuberculosis is complex. High-level resistance is associated with point mutations involving the ribosomal protein S12 (rpsL gene) and the S12-interacting regions of the 16S rRNA gene (rrs), i.e., in the proximity of positions 530 and 915 (3, 7, 17, 19). In Escherichia coli, ribosomal binding of KM is affected by mutations in the position 1400 region of the rrs gene (15).
Several investigations over 3 decades have shown that no cross-resistance occurs between SM and either AK or KM (10, 13, 23), but a general cross-resistance between AK and KM has repeatedly been demonstrated (1, 9, 18, 20, 26).
In Estonia, the incidence of primary resistance to any first-line drug among isolates from new pulmonary TB patients is more than 30% (5) and 13% of all culture-verified TB cases involve MDR disease. A majority of MDR M. tuberculosis isolates are resistant not only to rifampin and isoniazid but also to SM and ethambutol (6). Due to the high prevalence of drug resistance, there is a pronounced need for alternative agents and the deoxystreptamine aminoglycosides KM and AK are generally used in MDR-TB treatment. Even though it is generally believed that there is full cross-resistance in M. tuberculosis between these two drugs, we found susceptibility to AK in 43 of 79 (54%) KM-resistant clinical MDR-TB isolates from Estonian patients routinely tested in 2001 (12). The testing of these isolates for susceptibility to KM (4 μg/ml) and AK (1 μg/ml) was performed with the radiometric Bactec system at the Estonian National Reference Laboratory.
To further study this, we used sequencing of the rrs gene, DNA fingerprinting, and MIC determination for analyzing Estonian drug-resistant M. tuberculosis isolates. A total of 49 isolates were included in the study: 40 KM-resistant and AK-susceptible isolates, 4 isolates resistant to both drugs, and 5 dual-susceptible isolates, all from different patients.
MICs were determined in Middlebrook 7H10 agar supplemented with oleic acid-albumin-dextrose-catalase and 2 to 256 μg of KM or AK/ml. Resistance was defined as a MIC of >4 μg/ml (9, 16). All isolates were examined for mutations in the region of the rrs gene where substitutions giving resistance to KM-AK have been reported (1, 21). We amplified and sequenced an approximately 350-bp segment of the 16S rRNA gene, using the rrs.PCR.F123 (AAGGGCTGCGATGCCGCGAG) and rrs.PCR.R535 (AAGTCCGAGTGTTGCCTCAGG) primers. The PCR was carried out for 30 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s with AmpliTaq Gold (Applied Biosystems). PCR products were purified with a GFX PCR purification kit (Amersham Pharmacia Biotech) and sequenced in both directions with a Big Dye DNA sequencing kit (Applied Biosystems) The reaction mixtures were precipitated with ethanol and analyzed in an ABI Prism 3100 genetic analyzer (Applied Biosystems). Ten isolates were selected for additional sequencing of the complete rrs gene. All obtained sequences were compared, and substitution positions were numbered according to the CDC1551 public rrs gene (accession number AE 007009).
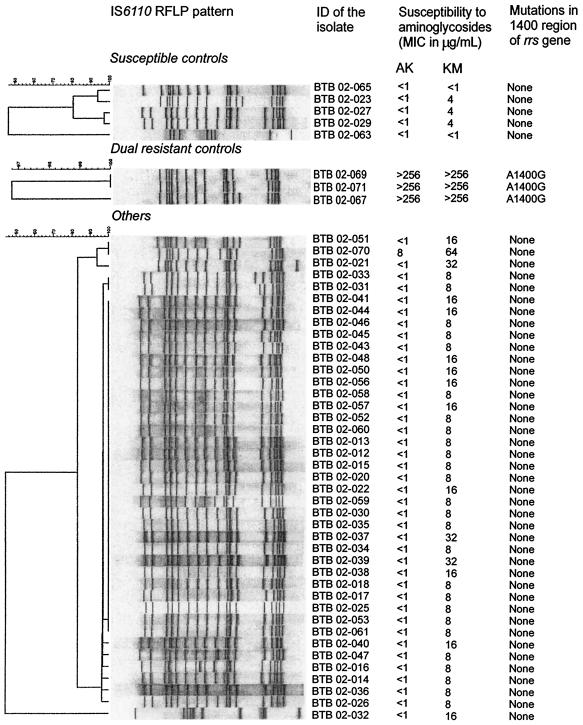
The M. tuberculosis isolates were genotyped by restriction fragment length polymorphism analysis, by using a standardized Southern blot hybridization method based on the insertion sequence IS6110 (24). The gels were scanned, and results were analyzed using the Gelcompar software (Applied Maths, Kortrijk, Belgium) as previously described (8). Although they were isolated from different patients, a high degree of similarity with a predominance of the Beijing genotype was seen among these MDR-TB isolates (Fig. 1).
FIG. 1.
Restriction fragment length polymorphism (RFLP) patterns and resistance to AK and KM of 49 isolates of M. tuberculosis originating from different patients and their mutations in the partially sequenced rrs gene. ID, identification number.
Forty KM-resistant isolates (MICs of 8 to 32 μg/ml) were confirmed susceptible to AK (MIC of ≤1 μg/ml). In one KM-resistant isolate (MIC of 64 μg/ml) low-level resistance to AK was detected (MIC of 8 μg/ml). For the three isolates included as dual-resistant controls, the MICs of both drugs were >256 μg/ml, while for the five susceptible controls the AK MICs were ≤1 and the KM MICs were 1 to 4 μg/ml.
All isolates highly resistant to both KM and AK revealed a guanine-for-adenine substitution at 16S rRNA position 1400 (Fig. 1). No mutations were seen in the position 1400 region of the rrs gene in isolates for which KM MICs were 64 μg/ml or less.
Ten M. tuberculosis isolates were further examined for mutations in the nucleotide sequence of the whole rrs gene. Besides confirmation of the A1400G mutation in one isolate, a thymine-for-cytosine substitution at 16S rRNA position 516 was detected in two additional isolates (MIC of KM, 32 to 64 μg/ml). No mutations in the rrs gene were identified in the remaining seven isolates (Table 1). To our knowledge the C516T mutation has not previously been associated with KM resistance in M. tuberculosis. However, in earlier reports the very same mutation has been found in SM-resistant strains and consequently suggested as an SM resistance marker (4, 7, 14). This, however, is in conflict with our findings, as our two isolates were KM resistant but SM susceptible.
TABLE 1.
Sequencing results of whole rrs gene analysis in 10 M. tuberculosis isolates having different resistance profiles for KM, AK, and SM
| Clinical isolate of M. tuberculosis | Susceptibility to aminoglycosidea
|
Mutation in the rrs gene | ||
|---|---|---|---|---|
| KM (MIC in μg/ml) | AK (MIC in μg/ml) | SMb | ||
| BTB 02-063 | S (1) | S (≤1) | S | None |
| BTB 02-065 | S (1) | S (≤1) | R | None |
| BTB 02-027 | S (4) | S (≤1) | R | None |
| BTB 02-012 | R (8) | S (≤1) | R | None |
| BTB 02-022 | R (16) | S (≤1) | R | None |
| BTB 02-021 | R (32) | S (≤1) | S | C516T |
| BTB 02-037 | R (32) | S (≤1) | R | None |
| BTB 02-039 | R (32) | S (≤1) | R | None |
| BTB 02-070 | R (64) | R (8) | S | C516T |
| BTB 02-067 | R (>256) | R (>256) | R | A1400G |
S, susceptible; R, resistant.
Drug concentration used, 4.0 μg/ml.
Victor et al. have proposed that the nucleotide change (C-to-T transition) at position 491 of the rrs gene (close to the position where we found the thymine-for-cytosine substitution) is a polymorphism not associated with drug resistance (25). These contradictory findings highlight the importance of establishing the causal relationship between any given mutation and drug resistance.
We did not find any mutations at positions 1400, 1401, and 1483 in any of the 40 KM-resistant and AK-susceptible MDR M. tuberculosis isolates tested. This is in agreement with earlier reports where no mutations were found in this region in low-level (MICs of ≤4 to 32 μg/ml) AK-KM-cross-resistant M. tuberculosis isolates (1). This and earlier studies suggest that nucleotide substitutions at position 1400 in the rrs gene may be used as an important marker of high-level AK-KM resistance (1, 21, 22). Since genetic methods have so far failed to detect all clinically relevant drug resistance to aminoglycosides, it is important to test antimicrobial susceptibilities of M. tuberculosis also by culture. Our data show that AK-KM cross-resistance is not generally present and indicate that there might be a need to test M. tuberculosis isolates with both these drugs.
Acknowledgments
This study was supported by the Swedish Baltic Sea Grant.
REFERENCES
- 1.Alangaden, G. J., B. N. Kreiswirth, A. Aouad, M. Khetarpal, F. R. Igno, E. K. Manavathu, and S. A. Lerner. 1998. Mechanism of resistance to amikacin and kanamycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayvazian, L. F. 1993. Tuberculosis—a comprehensive international approach, p. 1-20. In L. B. Reichman and E. S. Hershfield (ed.), History of tuberculosis. Marcel Dekker, Inc., New York, N.Y.
- 3.Böttger, E. C. 1994. Resistance to drugs targeting protein synthesis in mycobacteria. Trends Microbiol. 2:416-421. [DOI] [PubMed] [Google Scholar]
- 4.Dobner, P., G. Bretzel, S. Rüsch-Gerdes, K. Feldmann, M. Rifai, T. Löscher, and H. Rinder. 1997. Geographic variation of the predictive values of genomic mutations associated with streptomycin resistance in Mycobacterium tuberculosis. Mol. Cell. Probes 11:123-126. [DOI] [PubMed] [Google Scholar]
- 5.Espinal, M. A., A. Laszlo, L. Simonsen, F. Boulahbal, S. J. Kim, A. Reniero, S. Hoffner, H. L. Rieder, N. Binkin, C. Dye, R. Williams, and M. C. Raviglione. 2001. Global trends in resistance to antituberculosis drugs. N. Engl. J. Med. 344:1294-1303. [DOI] [PubMed] [Google Scholar]
- 6.Estonian Tuberculosis Registry. 2002. Tuberculosis incidence in Estonia 2000. Ühiselu AS, Tallinn, Estonia.
- 7.Finken, M., P. Kirschner, A. Meier, A. Wrede, and E. C. Böttger. 1993. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudo knot. Mol. Microbiol. 9:1239-1246. [DOI] [PubMed] [Google Scholar]
- 8.Heersma, H. F. K., K. Kremer, and J. D. A. van Embden. 1998. Computer analysis of IS6110 RFLP patterns of Mycobacterium tuberculosis. Methods Mol. Biol. 101:395-422. [DOI] [PubMed] [Google Scholar]
- 9.Heifets, L. B. 1991. Drug susceptibility tests in the management of chemotherapy of tuberculosis, p. 89-121. In L. B. Heifets (ed.), Drug susceptibility in the chemotherapy of mycobacterial infections. CRC Press, Inc., Boca Raton, Fla.
- 10.Hoffner, S. E., and G. Källenius. 1988. Susceptibility of streptomycin-resistant Mycobacterium tuberculosis strains to amikacin. Eur. J. Clin. Microbiol. Infect. Dis. 7:188-190. [DOI] [PubMed] [Google Scholar]
- 11.Iseman, M. D. 2000. Drug-resistant tuberculosis, p. 323-353. In M. D. Iseman (ed.), A clinician's guide to tuberculosis. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Krüüner, A. 2003. Drug-resistant Mycobacterium tuberculosis in Estonia. Ph.D. thesis. Karolinska Institute, Stockholm, Sweden.
- 13.McClatchy, J. K., W. Kanes, P. T. Davidson, and T. S. Moulding. 1977. Cross-resistance in M. tuberculosis to kanamycin, capreomycin, and viomycin. Tubercle 58:29-34. [DOI] [PubMed] [Google Scholar]
- 14.Meier, A., P. Sander, K. J. Schaper, M. Scholz, and E. C. Böttger. 1996. Correlation of molecular resistance mechanisms and phenotypic resistance levels in streptomycin-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 40:2452-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 16.Pfyffer, G. E., D. A. Bonato, A. Ebrahimzadeh, W. Gross, J. Hotaling, J. Kornblum, A. Laszlo, G. Roberts, M. Salfinger, F. Wittwer, and S. Siddiqi. 1999. Multicenter laboratory validation of susceptibility testing of Mycobacterium tuberculosis against classical second-line and newer antimicrobial drugs by using the radiometric BACTEC 460 technique and the proportion method with solid media. J. Clin. Microbiol. 37:3179-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 18.Riska, P. F., W. R. Jacobs, and D. Alland. 2000. Molecular determinants of drug resistance in tuberculosis. Int. J. Tuberc. Lung Dis. 4:S4-S10. [PubMed] [Google Scholar]
- 19.Sreevatsan, S., X. Pan, K. E. Stockbauer, D. L. Williams, B. N. Kreiswirth, and J. M. Musser. 1996. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob. Agents Chemother. 40:1024-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutton, W. B., R. S. Gordee, W. E. Wick, and L. V. Standfield. 1966. In vitro and in vivo laboratory studies on the antituberculous activity of capreomycin. Ann. N. Y. Acad. Sci. 135:947-959. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki, Y., C. Katsukawa, A. Tamaru, C. Abe, M. Makino, Y. Mizuguchi, and H. Taniguchi. 1998. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16 rRNA gene. J. Clin. Microbiol. 36:1220-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniguchi, H., B. Chang, C. Abe, Y. Nikaido, Y. Mizuguchi, and S. I. Yoshida. 1997. Molecular analysis of kanamycin and viomycin resistance in Mycobacterium smegmatis by use of the conjugation system. J. Bacteriol. 179:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukamura, M., and S. Mizuno. 1975. Cross-resistance relationships among the aminoglycoside antibiotics in Mycobacterium tuberculosis. J. Gen. Microbiol. 88:269-274. [DOI] [PubMed] [Google Scholar]
- 24.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. W. M. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Victor, T. C., A. van Rie, A. M. Jordaan, M. Richardson, G. D. van der Spuy, N. Beyers, P. D. van Helden, and R. Warren. 2001. Sequence polymorphism in the rrs gene of Mycobacterium tuberculosis is deeply rooted within an evolutionary clade and is not associated with streptomycin resistance. J. Clin. Microbiol. 39:4184-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization. 1997. Guidelines for the management of drug-resistant tuberculosis. WHO/TB/96.210. World Health Organization, Geneva, Switzerland.