Abstract
The completion of sequencing projects for various organisms has already advanced our insight into the evolution of entire genomes and the role of gene duplications. One multigene family that has served as a paradigm for the study of gene duplications and molecular evolution is the family of homeodomain-encoding genes. I present here an analysis of the homeodomain repertoire of an entire genome, that of the nematode Caenorhabditis elegans, in relation to our current knowledge of these genes in plants, arthropods, and mammals. A methodological framework is developed that proposes approaches for the analysis of homeodomain repertoires and multigene families in general.
Homeobox genes have been discovered in many species including animals, single-celled organisms such as yeast and dictyostelium, and plants. Intriguingly, there are often multiple duplicated versions of homeobox genes in vertebrates (best documented in mammals) as compared with the presence of just one homolog in the fruit fly Drosophila (1). This fact has been interpreted to mean that, during evolution, vertebrates developed more elaborate control mechanisms, presumably related to a more complicated body plan (2, 3). However, for some fly genes, e.g., bicoid (bcd), no vertebrate homologs could be identified despite substantial efforts. Similarly, many homeobox genes in the nematode Caenorhabditis elegans do not (yet?) have counterparts in the fly or mammalian genomes (4, 5).
This situation raises the possibility that different repertoires of homeobox genes may account for the complexity of regulatory controls, rather than simply gene number. Such a gene repertoire can be defined by several criteria: number of genes, types/classes of genes, variability among genes, and the diversity created. The latter aspects deserve special attention because the same spectrum of “sequence space” may be covered by few or many genes. For example, all major colors of the visual spectrum are represented in a box of 10 crayons, just as the same spectrum of colors would be covered by a box of 50 colored pencils. Similarly, the map of a landscape covers the same territory regardless of resolution, or, on a topological map, the distance of contour lines. Thus, greater resolution (denser contours, more genes) may not necessarily mean greater variability or diversity.
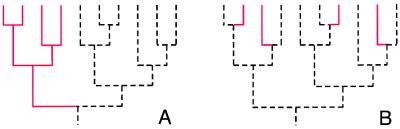
Applied to homeobox gene evolution, this concept implies that increasing complexity could have been accomplished by two quite different scenarios: (i) increased diversity (discovery of new territory, invention of “new” colors) through the evolution of classes of genes not present in other species; (ii) increased resolution (higher magnification, more topographical contour lines) through “fine” tuning of a repertoire that, in principle, overlaps with that of the evolutionary ancestors. Fig. 1 illustrates these two modes of generating increasing complexity: (i) acquisition of new major branches expanding the repertoire; and (ii) elaboration of existing major branches by “intercalation.”
Figure 1.
Models for the generation of homeodomain repertoires. Dashed lines represent branches and taxa shared with other species; solid lines represent gene acquisitions. (A) Scenario 1 postulates that gene acquisitions were associated with “novelty” so that the diversity of the prior repertoire was expanded in species-specific fashion by novel sequences and their subsequent duplications and divergence. (B) Scenario 2 postulates that the diversity of the repertoire is not fundamentally altered but is elaborated further by the creation of new branches as “intercalations” within the existing repertoire.
Although a definitive distinction between these two alternatives depends on evidence generated in the completion of multiple genome projects, the information available from C. elegans allows us already to operationalize the initial propositions. In this regard, the present study also serves to develop methodology for the investigation of multigene families and gene repertoires.
Materials and Methods
Homeodomain Sequences.
Amino acid sequences of homeodomains were collated either from the literature or from GenBank, Flybase, and Wormbase searches. Partial sequences were eliminated, and identical sequences were assumed to represent the same gene unless published information indicated otherwise. The compilation of datasets can be obtained as supplemental information from the PNAS web site (www.pnas.org).
Classification of Sequences.
Sequences were grouped into classes according to criteria established previously by using distance and cladistic methods (6). The inclusion of sequences in the Nkx-like and Prd-like classes in mammals was also in accordance with other classifications (1, 5).
Specific Data Subsets.
Each of the 80 C. elegans homeodomain sequences was classified into the subset “shared” or “unique,” respectively, depending on whether there are identifiable counterparts or orthologs in mammals. For this classification, I relied specifically on the analysis by Ruvkun and Hobert (5), who recently published a phylogenetic tree for C. elegans homeodomain sequences (see also www.sciencemag.org/feature/data/c-elegans.shl). Details on criteria for the classification of C. elegans sequences can be obtained from the PNAS web site.
Variability Plots.
The occurrence of specific amino acids for each position of the homeodomain sequence was determined by the character status function of paup 4.0 (Phylogenetic Analysis Using Parsimony, D. Swofford, from Sinauer Associates). Each residue was counted as one unit, and the number of units was plotted against the sequence position.
Distance Matrices and Distributions.
Simple distance matrices for various datasets were generated by the “Pairwise Distances” function of paup (6). For each distance length, the number of times present in the matrix was determined by using “Word Count” in Microsoft word 5.1a. The occurrence was plotted against the length of distance by using Microsoft excel 4.0. For control, homeodomain sequences were assembled, each of which was more than 90% dissimilar from the others (see www.pnas.org). Distance matrices were created by using artificial datasets of 10, 20, and 30 sequences. The distribution of distances for such highly nonrelated homeodomains was assumed to represent the hypothetical upper limit for analysis. This approach used existing sequences and avoided the need to apply structural criteria for assessing the contribution of residues to protein-folding capabilities of hypothetical molecules.
As datasets contained different numbers of sequences, the total number of data varied, resulting in different peak height for each curve. To allow for better visual comparisons, the curves in Figs. 4, 5 A, C, and E, and 6 B and D were adjusted along the y axis, so that each respective peak was set to 100%. The general shape of the curves remained unaltered.
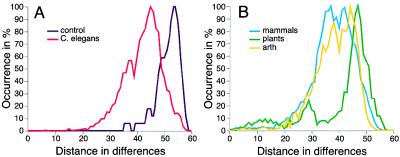
Figure 4.
Distribution of distances in pairwise comparisons. The total number of occurrences for every distance within a distance matrix was determined and then plotted for each dataset. For better comparison, all curves were adjusted in dimension along the y axis to the 100% level. A shows in purple the distribution curve derived from an artificial dataset of 30 most divergent homeodomain sequences. This curve represents the maximum possible distances observable from existing sequences. The C. elegans distance distribution curve is shown in red. Closer relationships between homeodomains result in an extension of the curve to the left of the graph. However, the majority of sequences are more than 50% dissimilar (>30 differences in 60 residues). B compares the distance distribution curves for mammalian (blue), plant (green), and Drosophila (yellow) sequences.
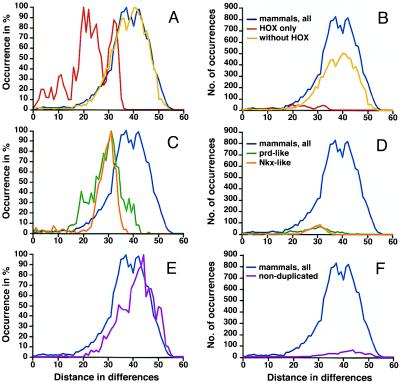
Figure 5.
Distance distributions for subets of mammalian homeodomains. A, C, and E show curves adjusted to 100% for the y axis; B, D, and F depict the actual distance counts. The overall shape of curves is identical between both graphs, which differ only with reference to the y axis. (A and B) Results for all distinct mammalian homeodomain sequences combined are shown in blue, results for HOX homeodomains in red, and results for the remaining sequences in yellow. (C and D) Distance distributions for Nkx-like (orange) and prd-like (green) homeodomains are compared with those for all mammalian sequences (blue). (E and F) Results for a mammalian sequence dataset from which all duplicates were removed. In comparison to the full mammalian sequences dataset, the curve peaks at a greater distance, and the shoulder indicative of related genes is markedly reduced.
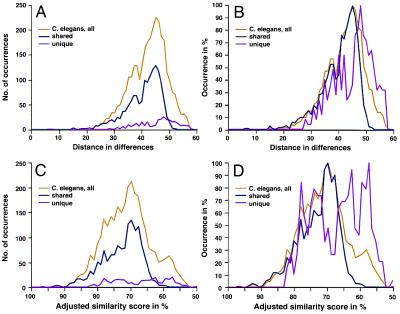
Figure 6.
Distance distributions for unique and evolutionarily conserved C. elegans homeodomains. Two distance measures were used: simple distance matrices that measure the number of differences over 60 positions (A and B) and the adjusted similarity scores (C and D), resulting from pairwise comparisons in which character state transitions were weighted according to a pam250 matrix (7). The absolute numbers of occurrences were determined separately (A and C), and curves adjusted to 100% are shown in B and D. Although the unique sequence dataset produces greater distances (pink) compared with the shared sequences (purple), it also indicates the presence of more closely related sequences, as evident by a second peak in the curve. A “shoulder” indicating closer relationships for a small number of sequences is also evident in the shared sequence dataset and the combined dataset (yellow).
Matrices with character weighting in accordance with the pam 250 dataset (7) were produced by using the align module of McMolly (Softgene, Berlin) and are expressed as adjusted similarity scores on a scale up to 100% (identity). For each similarity score, the occurrence was determined and plotted as described above.
Results
The objective of this work was to analyze a complete animal genome with respect to the repertoire of its homeobox genes. The underlying assumption was that the full homeobox gene complement of a species would allow a detailed understanding of the relationships of these genes and enable conclusions about their mode of evolution. With the completion of the C. elegans sequencing project, this genome is the first of a multicellular organism to become available for such analyses. Eventually, the hypotheses developed here will be testable by using comparisons of multiple complete genomes. In this regard, the present study also serves to develop methodology for such investigations.
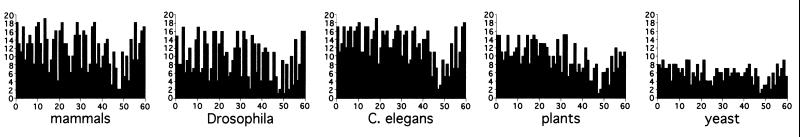
In a first approach, I estimated the variability of homeobox sequences in five major evolutionary phyla: C. elegans, Drosophila, mammals, yeast, and plants. The results are shown in Fig. 2. Interestingly, the extent of variability of amino acid residues in yeast homeodomain sequences (which represent 10 distinct classes) is similar to that found in Drosophila (72 distinct genes to date) or plants (70 genes to date). There is no appreciable increase in variability in mammals, which, with 175 distinct genes, harbor at least twice the number of sequences of the fly. The variability plot for C. elegans with 80 distinct genes is comparable to those for the other phyla. Critical inspection of the types of amino acids and characteristics of their side chains did not reveal gross differences in the appearance of specific residues at a given position (data not shown). These data indicate that the degree of variability in homeodomain sequences is not proportionally related to gene number.
Figure 2.
Variability plots for homeodomains in different clades. The occurrence of different amino acid residues at each position was plotted for mammals (175 sequences that represent distinct genes), Drosophila (72 distinct sequences), C. elegans (80 distinct sequences), plants (70 sequences), and yeast (10 distinct sequences). The most conserved positions are positions 48 and 49, which belong to the core residues of the DNA-binding helix. With the exception of the low number of amino acids observable in yeast, the variability plots are very similar for metazoans and plants.
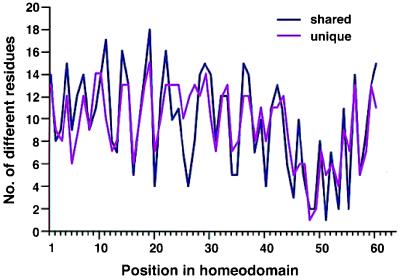
It is conceivable that the similarity of the C. elegans variability plot to that of mammalian sequences was solely a reflection of the fact that many gene classes are shared between the two clades. To test this, I assessed variability for C. elegans genes that have homologs in mammals (55 C. elegans sequences) and compared the results to those for C. elegans genes that do not (so far) have mammalian counterparts (25 C. elegans sequences). As shown in Fig. 3, the plots for both subgroups are comparable and vary by five or more residues only for positions 11, 26, 27, and 55. For residue 11, the unique sequences exhibit less variability, whereas for the other three, they exhibit higher variability. In contrast, position 26 is of low variability in mammals or arthropods, suggesting that these unique C. elegans homeodomains are indeed distinct. In general, however, the subset of unique genes, with about half as many sequences, produces the same variability as the subset of genes shared with mammals. Thus, the two groups of sequences exhibit the same diversity. This, again, supports the notion that diversity within a repertoire is not proportional to the number of sequences. The results also suggest that, should mammalian genes corresponding to (presently) unique C. elegans sequences be found, they would likely not increase the variability within the mammalian sequence repertoire. Taken together, these data provide evidence that (i) the overall diversity/variability in homeodomain sequences is not proportionally related to the number of sequences, and that (ii) the similarity in variability plots is not simply a reflection of corresponding/orthologous or homologous sequences in datasets. I conclude that diversity in the repertoire of homeodomains within a given species/clade is constituted more by the distance/dissimilarities of sequences than by gene number. This would imply, provided variability in two organisms is essentially similar, that the average distance between homeodomains should be larger in an organism with fewer genes and relatively smaller in an organism with a greater number of genes.
Figure 3.
Variability plots for evolutionarily conserved and unique C. elegans homeodomains. Conserved sequences are defined as “shared” with mammals (purple). Sequences so far unique to C. elegans were defined as “unique” (pink). The variability patterns for both datasets are very similar.
To test this prediction, I determined the distribution of distances from simple distance matrices that included all pairwise comparisons of distinct genes. To control for the potential influence of size of the dataset and to establish a baseline for the maximum possible extent of distances, I constructed artificial datasets of sequences from any organism that were most different. The results of control calculations (Fig. 4A) established the upper limit of maximum distances between two homeodomains. The peak of the distribution is at 53 differences within 60 positions, consistent with the high conservation of four to five residues within the DNA-binding helix of homeodomains (1). The prediction for actual species datasets is that the distribution would be shifted toward smaller distances (Fig. 4A, left in the graph) with the presence of more highly related sequences.
Indeed, the curve for the C. elegans dataset is located in the more modest distance range with the peak at 46 differences (in 60 positions). These data indicate the presence of some sequences in C. elegans that are more closely related, such as ceh-30 and ceh-31, for example. This is also reflected in a shoulder peak at 37, 38. Thus, the C. elegans homeodomain repertoire contains genes that may be recent duplicates of each other. For plant homeodomains, the distance curve is shifted toward greater distances with a peak at 47, but also has a much smaller second peak at 28. Clearly, plant genomes contain highly divergent homeodomains. Here, the second peak could result from the fact that the dataset contains related sequences from different plant species for which orthology had not been clearly established. Should these be distinct genes in different plant genomes, the second peak suggests a possible origin from more recent duplications. It should be kept in mind that only a fraction of the many plant homeodomains may have been identified to date.
The distance distributions for Drosophila and mammalian homeodomains each have two major peaks. The similarity in shape (despite a two-times greater number of sequences in mammals) confirms the above conclusion that repertoire diversity is not correlated with gene number. The maximum of the curve for fly homeodomain distances is found at 44 with a major second peak at 38. For the mammalian homeodomains, one major peak appears at 42, the second at 37. These data are consistent with the known presence of many subgroups of duplicates in the mammalian genome that are expected to shift the distribution curve to the left (Fig. 4A). Intermediate or short distances would be expected from such subgroups as the HOX/HOM class or prd-related homeodomains. Even for C. elegans, the extension of the curve to the left and the presence of a second peak at 37, 38 indicates the presence of related sequences. The differences in peak heights at 37, 38 between fly/mammals and C. elegans curves, however, attest that the overall fraction of duplicates is significantly smaller in the C. elegans repertoire.
To ascertain that multiple peaks in the distance distribution curves indeed reflect gene duplications, I analyzed the contribution of homeodomains known to have arisen by duplication in mammals (Fig. 5). The curve for HOX distances in itself has multiple peaks (at 32, 25, 22, and 20), which are also produced with only one sequence per paralogous group (HOXD13-HOXD9 and HOXB8-HOXB1; data not shown). As evident in Fig. 5A, HOX sequences are much more related to each other than the remaining sequences, and their shorter distances from each other contribute to the left peak of the mammalian distances curve. However, given their relatively minor fraction of the dataset (distances from comparisons of HOX genes amount to only 4.9% of the total distances; see Fig. 5B), these distances alone do not explain the shift of the highest peak to the left. Therefore, I also analyzed the contribution of other classes of duplicated genes; the Nkx-like and the prd-like sequences constitute the largest such groups. Fig. 5C shows that the distance distributions for both groups produce curves with one major peak at 31. This is close to the peak farthest to the right for the HOX class, indicating similar divergence for all three subgroups and an absence of closer duplicates in the Nkx-like class. From these results, it can be predicted that the peak of the curve for homeodomains that do not have duplicates would be positioned farther to the right, at greater distance. This is indeed the case: when the dataset consisted of only one sequence per class, the peak appears at 44 (Fig. 5E), very similar to the C. elegans curve in maximum position and shape. Thus, peaks to the left of distance maxima are produced by homeodomains that likely arose by gene duplication. It is noteworthy that the maximum divergence of sequences within a given mammalian subgroup is not different for the HOX, Nkx-like, and prd-like subclasses despite the fact that proteins with a prd-like homeodomain contain an additional conserved domain, the prd-domain (1). The data show that homeodomains in multidomain and homeodomain-only proteins are diverged similarly and suggest further that the different domains may be subject to independent evolutionary selection.
These analyses illustrate that the repertoire of homeodomains in C. elegans is more similar to that of arthropods and mammals than to plants. However, similarities in shapes of curves could reflect simply the conservation of homeodomain genes between animal genomes. To investigate the influence of conservation, I analyzed independently the variability and diversity for the subset of homeodomains that is shared between C. elegans and mammals and for those that are unique to C. elegans. Fig. 6A shows the curves for numbers of distances, and Fig. 6B depicts the curves adjusted to the 100% level. The homeodomains unique to C. elegans consist of two subgroups, as evidenced by the major peak at 48 and the cluster of smaller peaks (at 42, 40, 38, and 35) in the distribution curve. The highly dissimilar sequences (represented by distances peaking at 48) are as different from each other as those in plants (maximum at 47), whereas the minor peaks overlap with those for fly and non-HOX mammalian distances. These data indicate that the unique sequences are still notably divergent from each other. Thus, it is unlikely that they arose through very recent duplications (or potential subsequent gene losses) that occurred specifically only in the lineage leading to C. elegans. An almost identical pattern emerges when, instead of simple distances, weighted character state transitions are used. In this case, the specific residues occurring in each position and each pairwise comparison are weighted according to a pam250 matrix (7). As shown in Fig. 6 C and D, the peak for all C. elegans sequences coincides with that for sequences that are shared with mammals. In contrast, the curve for (so far) unique C. elegans homeodomains is shifted to the right (lower similarity scores) and has several peaks, with two major peaks in the area of lower similarity. These data are fully consistent with and validate the results derived from simple distance matrices (8) and provide strong support for the earlier conclusions. It should be noted here, nevertheless, that this analysis is preliminary in that the classification into shared and unique genes is based on current incomplete knowledge of the mammalian genomes. A more refined analysis of this hypothesis will become possible once the presence or absence of respective orthologs in a complete mammalian genome has been determined.
In taking these considerations together, I conclude that the diversity of homeodomain repertoires in different phyla is determined not by the overall number of genes present but rather by the relative distances between them. This outcome was ascertained by three measures of variability: (i) amino acid occurrence within homeodomain sequences, (ii) distributions of simple distances, and (iii) distributions of weighted similarity scores. In other words, fewer genes may produce the same variation as more genes so long as the overall spread/diversity is similar. This situation is illustrated further by the analysis of sequence subgroups within the mammalian homeodomain repertoire. Thus, there exists a good correlation between average sequence distance and diversity of the repertoire.
Although this result is not necessarily surprising, it has a number of important evolutionary implications: (i) An increased number of genes may provide greater complexity but does not automatically imply greater diversity of the repertoire. (ii) The mammalian lineage appears to have elaborated, by multiple gene duplications, a basic homeodomain repertoire that was shared with ancestors to arthropods and nematodes. Even although the mammalian and fly genomes are yet incomplete, they attest to more frequent gene duplications than in C. elegans. Clearly, there are only few examples of homeodomain duplicates in C. elegans, and it is unclear whether there were recent gene duplications. Nevertheless, the basic repertoire in mammals, and presumably the gene regulatory control mechanisms encoded by it, evolved fundamentally similarly to their invertebrate counterparts. (iii) The extent of diversity in homeodomain repertoires appears to be comparable for nematodes, arthropods, and mammals. Thus, the repertoires in different clades did not expand appreciably in variability during evolution. There are two possible explanations for this constancy: either the homeodomain possesses only limited “mutability” (presumably for structural reasons) or the time elapsed did not allow yet for greater divergence. The latter possibility is unlikely as some sequences exist that have evolved independently (such as for example, Drosophila bicoid and some genes unique to C. elegans). (iv) The evolutionary progression, at least for homeobox genes, more likely involved numerous duplications of terminal branches rather than invention of novelty. With respect to the hypotheses set forth at the start of this study, this favors model 2 (Fig. 1B) as the evolutionary scenario.
Discussion
In this work, I have analyzed the diversity in homeodomain repertoires in distinct evolutionary clades: plants and metazoans, comprising the nematode C. elegans, Drosophila as a representative of the arthropods, and mammals as the vertebrate prototype. My goal was to develop methodology and a theoretical framework that can be applied to homeodomain repertoires in many different genomes and that may serve ultimately as a paradigm for the study of multigene families, gene repertoires, and genome evolution.
Methodological Considerations: Limitations and Possible Future Approaches.
The first approach was to assess sequence diversity by variability plots. This method has two major advantages: it is simple and it does not make assumptions about evolutionary relationships. There are, however, several limitations: (i) The tabulation of residues at given positions of a sequence does not account for the possibility that a particular amino acid may be more likely at certain positions. More sophisticated frequency determinations could evaluate the “mutability” of a position (9). In this way, it would be possible to determine the relative contribution of a sequence position to evolutionary drift/diversification. (ii) The influence of neighboring residues is neglected. It is possible that combinations of amino acids are more likely to occur in homeodomains that belong to a special subclass, such as the paired class. To evaluate sequence diversity more precisely within a subclass, it is necessary to perform cluster analyses for neighboring residues or those in close spatial proximity (10). This approach would allow a more precise definition of the critical steps in the generation of homeodomain subclasses. Furthermore, such analyses would reveal concomitant changes in several residues (11) that could indicate a requirement for coevolution. (iii) Another measure of variability/diversity would be to devise strategies that measure the likelihood of occurrence of a particular actual gene out of the large number of hypothetically possible sequences. Multiplying the numbers of different residues found at each position in C. elegans homeodomains, for example, results in 1.32 × 1064 possible combinations if all residues are independent. With use of knowledge generated under ii or criteria that eliminate structurally impossible sequences, this number would become smaller. Nevertheless, the actual number of 80 homeodomains in the C. elegans genome is an exceedingly small fraction of the hypothetical possibilities. Obviously, there were specific selections made during evolution, and with some measure of the directions in which an extant homeodomain could have evolved, computational analyses would allow us to develop estimates about accompanying evolutionary time spans (12).
The second approach was to assess diversity by determining the distribution of pairwise distances between sequences. (iv) Major advantages of this approach are its simplicity and independence from underlying assumptions about evolutionary relationships (6, 8, 12). Disadvantages of distance approaches have been discussed extensively in the literature. With regard to homeodomain repertoires, I wish to address the following limitations: (v) In the absence of a calibration dataset, it is difficult to control variables that may influence the results, such as the number of sequences in a dataset. By creating an artificial dataset of selected divergent sequences, I was able to develop at least an estimate of the potential upper limit of results. However, the lower limits of resolution remain to be determined. Further, simple distance methods do not consider biological consequences of conservative or drastic changes. In the interest of relying on as few assumptions as possible, I did not introduce weighting parameters or significance measures to specific sequence differences. However, even when the pairwise comparisons of homeodomain sequences are weighted according to a pam250 matrix, the overall outcome of the analysis for homeodomain repertoires is highly similar (8) or, as in the case of C. elegans, the same (see Fig. 6). (vi) As duplicated genes are present in a given dataset, the resulting distribution curves tend toward smaller distances. This means that a distance approach most likely will not be suitable for comparative analysis of repertoires with similar fractions of duplicated genes, such as human and mouse. The curves would be similar even when the duplications occurred in completely distinct subfamilies (see Fig. 5); this is a particular limitation within the vertebrate lineage. (vii) It is well established that distance approaches underestimate evolutionary time in the absence of relative rate tests. For such tests to be feasible, a probable ancestor for any pair of sequences would have to be constructed. Similarly, the possibilities of multiple hits on the same site can be taken into account only when ancestors are known. (viii) It is generally believed that all homeodomains originated from an “Ur”-homeodomain (a great-grand ancestor; ref. 13), the identity of which remains to be determined. Then, the evolution of multiple homeodomain classes must be visualized less in linear models (such as in Fig. 1) but in a multidimensional sequence space. This space is defined by the totality of all randomly possible 60 residues of homeodomain sequences (20 amino acids at each of 60 positions = 1.15 × 1078 sequences) minus those that are structurally impossible. This completely hypothetical space is much larger than the possibilities mentioned under iii in the above paragraph, because that estimate was based on actual occurrence of residues in existing genes. Nevertheless, the relevance of comparing actual gene repertoires to the hypothetically possible space becomes obvious from the results such an approach can generate: it enables a quantitative measure of probability for actual genes and indicates how representative a given repertoire is. The trajectory that a given gene or group of genes has taken in this space can be mapped, and the variations while a gene evolved to its extant position can be simulated. A prediction from my results is that the space (including trajectories) used by repertoires with fewer genes is actually greater than that covered by repertoires composed of more genes. Third, quantitative estimates can be derived for expansion or contraction of gene subclasses. Fourth, in consideration of structural constraints, one could estimate how likely other evolutionary scenarios are (travel into a different direction in space). Indeed, the largely distinct repertoires of plant and vertebrate homeobox genes (8) suggest that scenarios of parallel evolution could be simulated on the basis of existing data. (ix) Although it is not possible currently to determine whether the extant homeodomains are most optimal in DNA-binding function, such information will become available from structural and mutagenesis approaches. Thus, the evolutionary selection on homeodomains appears to involve properties beyond DNA binding. For RNA structures, it is now possible to assign relative functionality values to any real or hypothetical RNA sequence (14, 15). This enables so-called “walks on landscapes” in which correlations are drawn between local optima and the existence of specific RNA structures. I envision analogous walks through homeodomain sequence space or on homeodomain landscapes (16). Such an approach would describe the evolution of this multigene family not only by sequence comparisons but also in functional terms, relating their DNA-binding and gene regulatory capabilities. (x) Lastly, the ability to design optimal homeodomains computationally on the basis of methods outlined above will enable us to simulate future evolutionary trajectories. Sequence analysis approaches that incorporate structural and functional considerations (17) will thus not only provide novel insights into the evolution of multigene families but will also constitute a novel framework for the analysis of gene repertoires in complete genomes.
Implications for the Evolution of Homeodomain Repertoires.
Some important conclusions can be drawn from the analyses I present here: (i) If complexity of the homeodomain repertoire is defined by gene number and diversity, my results enable an assessment of the relative contribution of these two parameters. The comparisons of the C. elegans repertoire with other repertoires indicate that gene number alone is not sufficient to increase diversity. Rather, the relative distinctness of homeodomains within a species defines diversity. Apparently, despite increasing gene number, mammals may not have evolved a more diverse repertoire. (ii) The larger gene number in mammals, as accomplished by gene duplications, means that new functions for duplicates could be developed in two ways: through modification/mutation of the homeodomain so that new DNA-binding specificities or new protein interactions are acquired, or through changes in the regulation of expression of each gene (3, 18). The latter notion underscores the importance of upstream gene regulatory mechanisms that may place constraints on homeodomain diversification: in the case of the mammalian HOX genes, their organization in clusters contributes to regulation of gene expression and thus constrains divergence by genomic mechanisms rather than at the protein level (19–22). Further, the transcriptional activity of HOX clusters is controlled by regulators that are evolutionarily conserved, such as the polycomb and homothorax group genes (for review, see ref. 23). It is conceivable that networks of regulatory controls, rather than individual homeodomains, have been subject to selection and conservation. (iii) It has often been suggested that on gene duplication, one copy becomes “frozen” in sequence and/or function, whereas the other is free to diverge (24). For homeodomains, this possibility is best analyzed for the classical HOX genes, as comparisons can be done in parallel for each paralogous group of genes. There is no convincing evidence that “freezing” of a cluster or individual gene was a prominent mode in the evolution of the HOX genes (25). Rather, HOX sequences on all four clusters are similarly diverged, making it difficult even to define an ancestral cluster (26). It is more appealing to apply the “freeze” hypothesis to entire repertoires of homeodomains, at least in metazoans. The high degree of conservation between C. elegans, Drosophila, and mammals suggests that, once invented in an early metazoan ancestor, the repertoire was fine-tuned by “intercalation” rather than continuously expanded. Thus, if at all, constancy was imposed on an entire repertoire rather than on specific gene copies. This proposition is underscored by the striking conservation in regulation of expression for homeobox-containing genes across vertebrates (27–29). If there were different selective pressures on original genes and duplicates, some variation would be expected between distant vertebrate lineages. (iv) Finally, the homeodomain repertoire of C. elegans does not provide evidence that different homeodomains have evolved at entirely different rates within this species. This would be the only way to gain nematode-specific homeodomains significantly different from those in other animals. The results of this study, however, favor the evolutionary model in Fig. 1B (scenario 2). The model entails the elaboration of an early homeodomain repertoire by gene duplications and “intercalation” of the new genes into the existing repertoire. Evolution of “novelty” was accomplished within existing boundaries of diversity rather than by expanding the boundaries of the repertoire itself. Thus, the major evolutionary innovation was the establishment of homeodomains as regulatory motifs and of a network of developmental regulatory controls. On this basis, evolution then proceeded with modifications and elaboration of diversity within limits. As more complex analysis tools become available and genome projects are completed, these hypotheses can be evaluated further for the homeodomain gene family and may provide a general theoretical framework for study of the evolution of multigene families.
Supplementary Material
Acknowledgments
I acknowledge El Camino Del Diablo for inspiration and express gratitude to Dr. J. Michael Salbaum (Department of Cell Biology and Anatomy, Center for Human Molecular Genetics, Munroe–Meyer Institute, University of Nebraska Medical Center) for continuing critical discussion. I thank two anonymous reviewers for stimulating comments. Research in my laboratory was partially supported by the Mayo Foundation for Medical Education and Research.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bürglin T R. In: The Evolution of Homeobox Genes. Arai R, Kato M, Doi Y, editors. Tokyo: The National Science Museum Foundation; 1995. pp. 291–336. [Google Scholar]
- 2.Holland P W, Garcia-Fernandez J, Williams N A, Sidow A. Development (Cambridge, U.K.) 1994. S125–S133. [PubMed] [Google Scholar]
- 3.Ruddle F H, Bartels J L, Bentley K L, Kappen C, Murtha M T, Pendleton J W. Annu Rev Genet. 1994;28:423–442. doi: 10.1146/annurev.ge.28.120194.002231. [DOI] [PubMed] [Google Scholar]
- 4.Sharkey M, Graba Y, Scott M P. Trends Genet. 1997;13:145–151. doi: 10.1016/s0168-9525(97)01096-2. [DOI] [PubMed] [Google Scholar]
- 5.Ruvkun G, Hobert O. Science. 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- 6.Kappen C, Schughart K, Ruddle F H. Genomics. 1993;18:54–70. doi: 10.1006/geno.1993.1426. [DOI] [PubMed] [Google Scholar]
- 7.Dayhoff M O, Barker W C, Hunt L T. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 8.Kappen C. Computers Chem. 2000;24:95–103. doi: 10.1016/s0097-8485(99)00049-2. [DOI] [PubMed] [Google Scholar]
- 9.Wilbur W J. Mol Biol Evol. 1985;2:434–437. doi: 10.1093/oxfordjournals.molbev.a040360. [DOI] [PubMed] [Google Scholar]
- 10.Tatusov R L, Koonin E V, Lipman D J. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 11.Feng D F, Cho G, Doolittle R F. Proc Natl Acad Sci USA. 1997;94:13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kappen C. Computers Chem. 1996;20:49–59. doi: 10.1016/s0097-8485(96)80007-6. [DOI] [PubMed] [Google Scholar]
- 13.Bürglin T R. Dev Genes Evol. 1998;208:113–116. doi: 10.1007/s004270050161. [DOI] [PubMed] [Google Scholar]
- 14.Cupal J, Flamm C, Renner A, Stadler P F. ISMB. 1997;5:88–91. [PubMed] [Google Scholar]
- 15.Huynen M A, Hogeweg P. J Mol Evol. 1994;39:71–79. doi: 10.1007/BF00178251. [DOI] [PubMed] [Google Scholar]
- 16.Fontana W, Schuster P. Biophys Chem. 1987;26:123–147. doi: 10.1016/0301-4622(87)80017-0. [DOI] [PubMed] [Google Scholar]
- 17.Tomandl D, Schober A, Schwienhorst A. J Comput Aided Mol Des. 1997;1997:29–38. doi: 10.1023/a:1008071310472. [DOI] [PubMed] [Google Scholar]
- 18.Kappen C, Ruddle F H. Curr Opin Genet Develop. 1993;3:931–938. doi: 10.1016/0959-437x(93)90016-i. [DOI] [PubMed] [Google Scholar]
- 19.Gerard M, Chen J-Y, Gronemeyer H, Chambon P, Duboule D, Zakany J. Genes Dev. 1996;10:2326–2334. doi: 10.1101/gad.10.18.2326. [DOI] [PubMed] [Google Scholar]
- 20.van der Hoeven F, Zakany J, Duboule D. Cell. 1996;85:1025–1035. doi: 10.1016/s0092-8674(00)81303-3. [DOI] [PubMed] [Google Scholar]
- 21.Kondo T, Duboule D. Cell. 1999;97:407–417. doi: 10.1016/s0092-8674(00)80749-7. [DOI] [PubMed] [Google Scholar]
- 22.Sharpe J, Nonchev S, Gould A, Whiting J, Krumlauf R. EMBO J. 1998;17:1788–1798. doi: 10.1093/emboj/17.6.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher A, Magnuson T. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 24.Ruddle F H, Bentley K L, Murtha M T, Risch N. Development (Cambridge, U.K.) 1994. S155–S161. [PubMed] [Google Scholar]
- 25.Kappen C. Computational Medicine, Public Health and Biotechnology: Building a Man in the Machine. Vol. 1 1995. , Ser. Math. Biol. Med., ed. Wiiten, M. 5, 211–233. [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey W J, Kim J, Wagner G P, Ruddle F H. Mol Biol Evol. 1997;14:843–853. doi: 10.1093/oxfordjournals.molbev.a025825. [DOI] [PubMed] [Google Scholar]
- 27.Shashikant C S, Kim C B, Borbely M A, Wang W C, Ruddle F H. Proc Natl Acad Sci USA. 1998;95:15446–15451. doi: 10.1073/pnas.95.26.15446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall H, Studer M, Pöpperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. Nature (London) 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- 29.Vesque C, Maconochie M, Nonchev S, Ariza-McNaughton L, Kuroiwa A, Charnay P, Krumlauf R. EMBO J. 1996;15:5383–5396. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.