Abstract
A biapenem-tolerant mutant of Pseudomonas aeruginosa was isolated by Tn1737KH insertion. The survival of the mutant 3 h after the addition of biapenem was about 1,000 times greater than that of the wild type. The mutant was also tolerant to other biapenems, such as imipenem, panipenem, and meropenem.
Pseudomonas aeruginosa infections are difficult to eradicate with antibiotic treatment. It is well-known that biofilm bacteria are more resistant to the killing effect of antibiotics than planktonic bacteria. It has previously been reported that adherent bacteria on a solid surface were already tolerant to antibiotics even before biofilm formation (5). It seems that one of the mechanisms of tolerance is related to a stress response by adherent cells to attachment stimuli on a solid surface. In that case, an antibiotic-tolerant mutant(s) which cannot regulate the gene involved in survival during attachment on a solid surface will probably be obtained among the planktonic cells. We have searched for the gene(s) involved in bacterial survival in the presence of antibiotics and, as a result, isolated a mutant more resistant to the killing by biapenem than the wild type from planktonic cells of P. aeruginosa. Here we report a gene, termed tcp in this study, that has a suppressive role in antibiotic tolerance.
The bacterial strains, plasmids, and primers used in this study are listed in Table 1. DNA manipulations were performed by standard procedures (6). The MIC and minimal bactericidal concentration (MBC) of each antibiotic were determined by the broth microdilution method as previously described (5) with the following modification: the bacterial suspensions at a density of 0.5 × 106 to ∼1 × 106 cells/ml were incubated in Luria-Bertani (LB) broth.
TABLE 1.
Bacterial strains, plasmids and primers used in this study
| Strain, plasmid, or primer | Genotype, description, or sequence | Reference or source |
|---|---|---|
| Plasmids | ||
| pACΩGm | pACYC184 derivative carrying Ω fragment, Gmr | 7 |
| pMOB3 | Kmr Cmr, 5% sucrose sensitive | 8 |
| pGEM-T Easy | Apr and TA cloning vector | Promega |
| pMT6121 | oriT, sacB, and Hgr Kmr in Tn1737KH | M. Tsuda (14) |
| pTK01 | tcp in pGEM-T Easy | This study |
| pTKG01 | tcp::Gmr in pGEM-T Easy | This study |
| pTKM01 | tcp::Gmr and mob in pGEM-T Easy | This study |
| pCR2.1 | Apr Kmr and TA cloning vector | Invitrogen |
| pTKM02 | tcp and mob in pCR2.1 | This study |
| pTKM03 | tcp, orf2, and mob in pCR2.1 | This study |
| pTKM04 | tcp and mob in pCR2.1 | This study |
| Strains | ||
| P. aeruginosa | ||
| SM7 | Smr derivative of PAO1 | This study |
| SM21 | Smr derivative of PAO1 | This study |
| KMX78 | Kmr Hgr, SM7 derivative harboring pMT6121::Tn1737KH | This study |
| KMX7803 | SM7 tcp::Tn1737KH(Kmr Hgr) | This study |
| TKP011 | SM21 tcp::Gmr | This study |
| TKP02 | KMX7803 derivative harboring pTKM02 | This study |
| TKP03 | KMX7803 derivative harboring pTKM03 | This study |
| TKP04 | TKP011 derivative harboring pTKM04 | This study |
| E. coli | ||
| CT726 | DH1(pMT6121::Tn1737KH) | M. Tsuda (14) |
| S17-1 | thi endA recA hsdR with RP4-2Tc:: Mu-Km::Tn7 integrated in chromosome | 10 |
| Primers | ||
| galk-s | 5′-CGCAGAACAGGCAGCAGAGCGT TT-3′ | This study |
| galk-a | 5′-GCAGCAGAGGCGGATAAAAGT GCG-3′ | This study |
| PAO561-s1 | 5′-GTGAAGGAAAGCGAGCACAACA AGGCGT-3′ | This study |
| PAO561-a | 5′-CGGGGTTAGAATGCCTCGCTC CCA-3′ | This study |
| PAO561-s2 | 5′-GTGATTGCCACTGTCCCACTCCG CTCAT-3′ | This study |
Susceptibility to antibiotics was also determined by the killing test. The cultures were grown overnight in LB broth supplemented with the appropriate antibiotics, and the bacterial cells were resuspended in 10 ml of LB broth. The suspension was incubated for 4 to 10 h at 37°C in the presence of each carbapenem, at a concentration ranging from 8 to 32 μg/ml. The number of live cells was determined by colony counting.
The transposon insertion mutant(s) was constructed as follows. Escherichia coli strain CT726 harboring Tn1737KH in plasmid pMT6121 (14), which is a temperature-sensitive plasmid for replication, was mobilized to P. aeruginosa SM7 cells (9). The kanamycin- and mercury-resistant and temperature-sensitive transconjugant was isolated. The transconjugant P. aeruginosa KMX78 strain was grown at 30°C for 17 h, and then the cells were transferred to 96-well microtiter plates. The plates were incubated at 42°C for 24 h, and temperature-resistant mutants with the Tn1737KH insertion in their chromosome were selected. Strains harboring the chromosomal Tn1737KH insertion were first inoculated into LB broth supplemented with biapenem (8 μg/ml) in 96-well plates. Cultures that showed inhibited growth were collected from the wells and plated on LB agar containing kanamycin and mercury chloride.
The antibiotic susceptibility of strains obtained was determined by the broth microdilution method, and the biapenem-tolerant mutant P. aeruginosa KMX7803 strain was isolated. The Tn1737KH insertion site was determined by inverse PCR amplification using primers galk-s and galk-a and the Dye Terminator Cycle Sequence method. The Tn1737KH transposon had been inserted into the open reading frame (ORF) of 1,150 bp referred to as PA0561 (GenBank accession no. AE004492) in the Pseudomonas Genome Project. No gene showing high homology to the ORF was found. The gene corresponding to this ORF was named tcp (named tcp for tolerance to carbapenem in planktonic cells).
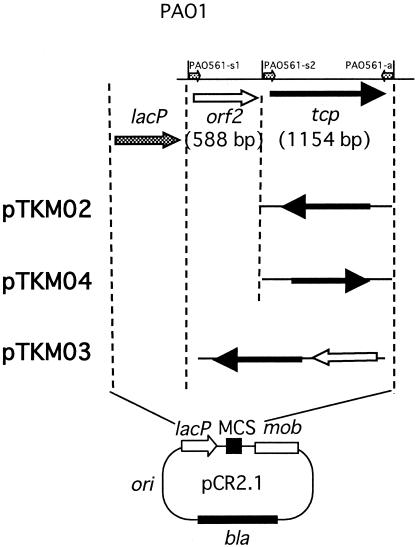
In order to obtain a tcp gene knockout mutant, first, we amplified the tcp gene by PCR using primers PAO561-s1 and PAO561-a (GenBank accession no. AE004492; protein identification no. AAG03950.1) and PAO1 DNA as a template. Plasmid pTK01 was constructed by insertion of the tcp gene into a pGEM-T Easy vector (Promega). The gentamicin resistance gene of pACΩGm (7) was then inserted into the HindIII site of pTK01 to form pTKG01. Plasmid pTKM01 was constructed by ligation of a NotI fragment containing tcp::Gmr from pTKG01 and a NotI fragment containing the mob cassette from pMOB3 (10), and it was transformed into E. coli S17-1 (10). After conjugal transfer of pTKM01 into P. aeruginosa SM21, we selected gentamicin-resistant strain TKP011, which contained the tcp::Gmr insertion in place of the tcp gene, and confirmed the insertion site by DNA sequencing analysis. For complementation testing, plasmids pTKM02, pTKM03, and pTKM04 were constructed (Fig. 1), and complemented P. aeruginosa TKP02, TKP03, and TKP04 strains were obtained.
FIG. 1.
Scheme showing the construction of plasmids for complementation testing. The primers ( 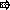 ) for PCR were as follows: primers PAO561-s2 and PAO561-a for the tcp structural region only and primers PAO561-s1 and PAO561-a for the tcp structural gene and the upstream region of orf2. Plasmid pTKM02 carried the tcp structural region in the orientation opposite that of the lac promoter in the pCR2.1 vector. Plasmid pTKM03 carried the tcp and the orf2 in the reverse direction, and pTKM04 carried the tcp structural gene in the forward direction. The tcp gene [
) for PCR were as follows: primers PAO561-s2 and PAO561-a for the tcp structural region only and primers PAO561-s1 and PAO561-a for the tcp structural gene and the upstream region of orf2. Plasmid pTKM02 carried the tcp structural region in the orientation opposite that of the lac promoter in the pCR2.1 vector. Plasmid pTKM03 carried the tcp and the orf2 in the reverse direction, and pTKM04 carried the tcp structural gene in the forward direction. The tcp gene [ 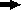 ], orf2 gene [
], orf2 gene [ 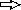 ], and lac promoter [
], and lac promoter [ 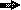 ] are shown. Drawings are not to scale. MCS, multiple cloning site.
] are shown. Drawings are not to scale. MCS, multiple cloning site.
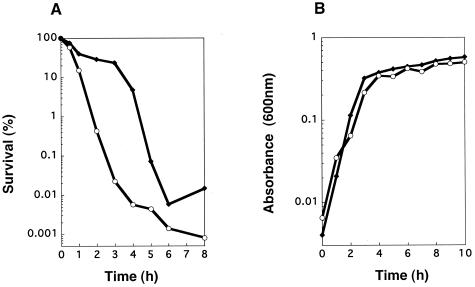
Whereas the MIC of biapenem for P. aeruginosa strain KMX7803 was exactly the same as the MIC for the wild type, the MBC for the strain was 16 times higher than that for the wild type (Table 2). The number of mutant cells 3 h after the addition of biapenem (32 μg/ml) was about 1,000 times greater than that of the wild type (Fig. 2A), while the growth curve for KMX7803 was the same as that for the wild type (Fig. 2B). MICs of cefoperazone, cefepime, gentamicin, and ofloxacin for KMX7803 were exactly the same as those for the wild type, except for the MIC of gentamicin, which was two times higher than the MIC for the wild type. The MBCs of cefoperazone, cefepime, gentamicin, and ofloxacin for KMX7803 were the same as those for the wild type (Table 2).
TABLE 2.
Antibiotic susceptibility of planktonic cells
| Strain | Biapenem
|
Cefoperazone
|
Cefepime
|
Ofloxacin
|
Gentamicin
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| MICa | MBCa | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| SM7b | 1 | 1 | 4 | 8 | 2 | 2 | 1 | 1 | 0.25 | 0.5 |
| KMX7803c | 1 | 16 | 4 | 8 | 2 | 2 | 1 | 1 | 0.5 | 0.5 |
MICs and MBCs given in micrograms per milliliter.
tcp+ wild-type strain.
SM7 tcp::Tn1737KH.
FIG. 2.
(A) Killing curves for biapenem (32 μg/ml) on P. aeruginosa SM7 and KMX7803 and (B) growth curves of P. aeruginosa SM7 and KMX7803. For panel A, assuming that the survival at zero time (1 × 109 to 2 × 109 CFU/ml) was 100%, the number of CFU was changed to a percentage. Symbols:  , SM7 (wild type);
, SM7 (wild type);  , KMX7803 (tcp::Tn1737KH).
, KMX7803 (tcp::Tn1737KH).
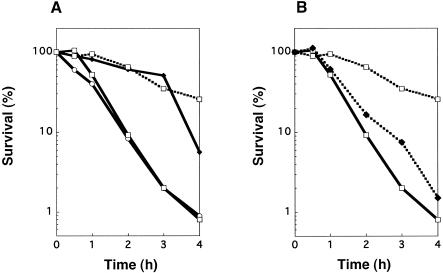
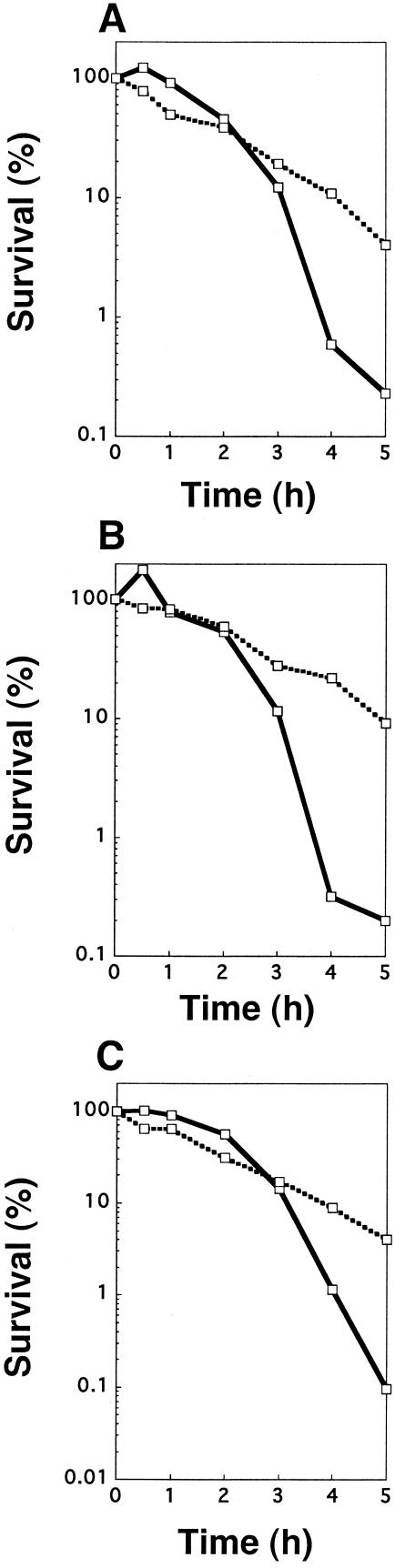
The MBC of biapenem for the P. aeruginosa tcp::Gmr strain TKP011 was 64 times higher than the MIC. The MBC/MIC ratios of the other carbapenems for the mutant were not as high as that of biapenem (Table 3). It appears that MBC is not a suitable or reliable measure of tolerance, because it is defined as the endpoint survival of more than 99.9% cells. Accordingly, cell viability in the presence of antibiotic was determined to clarify whether the tcp gene affects only biapenem susceptibility. The survival rate of P. aeruginosa TKP011 after incubation in the presence of biapenem at 8 μg/ml was as high as that of P. aeruginosa KMX7803 (Fig. 3A). The survival rates of TKP011 to imipenem, meropenem, and panipenem were 10 to 100 times higher than those of the wild type (Fig. 4). Moreover, the survival rate of TKP04, complemented strain harboring pTKM04 (Fig. 1), was as low as that of the wild type (Fig. 3B). The survival of another complemented strain, TKP03, was also equally low as that of the wild type (data not shown). In addition, the survival of TKP02, possibly lacking the tcp promoter, was shown to be as high as that of KMX7803 (data not shown). Therefore, constructed plasmids pTKM03 and pTKM04 could complement the mutation in strains KMX7803 and TKP011, respectively, but pTKM02 could not do so.
TABLE 3.
Carbapenem susceptibility of tcp knockout mutants, complemented strain, and parent strains
| Strain | Biapenem
|
Imipenem
|
Panipenem
|
Meropenem
|
||||
|---|---|---|---|---|---|---|---|---|
| MICa | MBCa | MIC | MBC | MIC | MBC | MIC | MBC | |
| SM7b | 1 | 1 | 1 | 2 | 8 | 16 | 1 | 2 |
| SM21b | 1 | 1 | 1 | 2 | 4 | 8 | 1 | 2 |
| KMX7803c | 1 | 16 | 2 | 4 | 8 | 16 | 1 | 2 |
| TKP011d | 0.125 | 8 | 2 | 4 | 4 | 16 | 0.5 | 4 |
| TKP04e | 0.5 | 4 | 4 | 16 | 8 | 16 | 4 | 4 |
MICs and MBCs given in micrograms per milliliter.
tcp+ wild-type strain.
SM7 tcp::Tn1737KH.
SM21 tcp::Gm.
TKP011(pTKM04 tcp+).
FIG. 3.
Killing curve of tcp mutants in the presence of biapenem at 8 μg/ml. Assuming that the survival at zero time was 100% (1 × 109 to 2 × 109 CFU/ml), the number of CFU was changed to a percentage. (A) Killing curve of tcp knockout mutant. [ ], P. aeruginosa SM7 (parent strain for KMX7803); [
], P. aeruginosa SM7 (parent strain for KMX7803); [ ], P. aeruginosa KMX7803 (tcp::Tn1737KH); [
], P. aeruginosa KMX7803 (tcp::Tn1737KH); [ ], P. aeruginosa SM21 (parent strain for TKP011); [▪ ▪ ▪ ▪□▪ ▪ ▪ ▪], P. aeruginosa TKP011 (tcp::Gmr) (B) Complementation of tcp gene. [
], P. aeruginosa SM21 (parent strain for TKP011); [▪ ▪ ▪ ▪□▪ ▪ ▪ ▪], P. aeruginosa TKP011 (tcp::Gmr) (B) Complementation of tcp gene. [ ], P. aeruginosa SM21; [▪ ▪ ▪ ▪□▪ ▪ ▪ ▪], P. aeruginosa TKP011 (tcp::Gmr); [▪ ▪ ▪ ▪⧫▪ ▪ ▪ ▪], P. aeruginosa TKP04 (tcp::Gmr, pTKM04).
], P. aeruginosa SM21; [▪ ▪ ▪ ▪□▪ ▪ ▪ ▪], P. aeruginosa TKP011 (tcp::Gmr); [▪ ▪ ▪ ▪⧫▪ ▪ ▪ ▪], P. aeruginosa TKP04 (tcp::Gmr, pTKM04).
FIG. 4.
Killing curves of P. aeruginosa SM21 and TKP011 in the presence of carbapenems. Assuming that the survival at zero time was 100% (1 × 109 to 2 × 109 CFU/ml), the number of CFU was changed to a percentage. Samples were incubated in the presence of imipenem at 16 μg/ml, panipenem at 8 μg/ml, or meropenem at 12 μg/ml for 5 h. After that, the number of CFU were counted. [ ], SM21; [▪ ▪ ▪ ▪□▪ ▪ ▪ ▪], TKP011.
], SM21; [▪ ▪ ▪ ▪□▪ ▪ ▪ ▪], TKP011.
The results of this study show that the number of live cells of the tcp mutant was higher than that of the wild-type strain in the presence of carbapenem. We found no known motif sequence of penicillin-binding proteins in the Tcp protein and no homology between penicillin-binding proteins and Tcp. Autolysis in tcp-defective mutants was not affected by the tcp gene (data not shown). However, it cannot be concluded that the tcp gene is entirely unrelated to the autolytic process in P. aeruginosa (12). Biofilm and stationary-phase cells are more tolerant than planktonic cells (1-4, 13, 16). Tolerance to antibiotics in stationary-phase or biofilm cultures is largely dependent on the presence of persister cells, according to Spoering and Lewis (11). After biapenem addition, survival of adherent cells of the tcp mutant on a plastic surface was higher than that of planktonic cells (preliminary data). We are not sure whether the tcp gene affects the biapenem tolerance of adherent cells of Pseudomonas aeruginosa. It was reported that viability of attached bacteria was very high and that physiological adaptations occurred during the early phases of attached growth (15). For full understanding of the role of the tcp gene in the mechanism of tolerance to antibiotics, further study is needed to identify the Tcp protein and to elucidate the regulation of the tcp gene.
Acknowledgments
We thank M. Tsuda for providing strain CT726 harboring Tn1737KH. We thank Herbert P. Schweizer for providing plasmids pMOB3 and pACΩGm.
This work was supported by a grant-in-aid for scientific research (no. 12671774) to T.O. from the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Aaron, S. D., W. Ferris, K. Ramotar, K. Vandemheen, F. Chan, and R. Saginur. 2002. Single and combination antibiotic susceptibilities of planktonic, adherent, and biofilm-grown Pseudomonas aeruginosa isolates cultured from sputa of adults with cystic fibrosis. J. Clin. Microbiol. 40:4172-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, D. G., and P. Gilbert. 1995. Modification by surface association of antimicrobial susceptibility of bacterial populations. J. Ind. Microbiol. 15:311-317. [DOI] [PubMed] [Google Scholar]
- 3.Charpentier, E., R. Novak, and E. Tuomanen. 2000. Regulation of growth inhibition at high temperature, autolysis, transformation and adherence in Streptococcus pneumoniae by ClpC. Mol. Microbiol. 37:717-726. [DOI] [PubMed] [Google Scholar]
- 4.Chuard, C., P. E. Vaudaux, R. A. Proctor, and D. P. Lew. 1997. Decreased susceptibility to antibiotic killing of a stable small colony variant of Staphylococcus aureus in fluid phase and on fibronectin-coated surfaces. J. Antimicrob. Chemother. 39:603-608. [DOI] [PubMed] [Google Scholar]
- 5.Miyake, Y., S. Fujiwara, T. Usui, and H. Suginaka. 1992. Simple method for measuring the antibiotic concentration required to kill adherent bacteria. Chemotherapy 38:286-290. [DOI] [PubMed] [Google Scholar]
- 6.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Schweizer, H. P. 1993. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 8.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 9.Simon, R., M. O'Connell, M. Labes, and A. Pühler. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 10.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 11.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomasz, A., A. Albino, and E. Zanati. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138-140. [DOI] [PubMed] [Google Scholar]
- 13.Trafny, E. A. 1998. Susceptibility of adherent organisms from Pseudomonas aeruginosa and Staphylococcus aureus strains isolated from burn wounds to antimicrobial agents. Int. J. Antimicrob. Agents 10:223-228. [DOI] [PubMed] [Google Scholar]
- 14.Tsuda, M. 1998. Use of a transposon-encoded site-specific resolution system for construction of large and defined deletion mutations in bacterial chromosome. Gene 207:33-41. [DOI] [PubMed] [Google Scholar]
- 15.Williams, I., F. Paul, D. Lloyd, R. Jepras, I. Critchley, M. Newman, J. Warrack, T. Giokarini, A. K. Jayes, P. F. Randerson, and W. A. Venables. 1999. Flow cytometry and other techniques show that Staphylococcus aureus undergoes significant physiological changes in the early stages of surface-attached culture. Microbiology 145:1325-1333. [DOI] [PubMed] [Google Scholar]
- 16.Williams, I., W. A. Venables, D. Lloyd, F. Paul, and I. Critchley. 1997. The effects of adherence to silicone surfaces on antibiotic susceptibility in Staphylococcus aureus. Microbiology 143:2407-2413. [DOI] [PubMed] [Google Scholar]