Abstract
We isolated mutant YM644, which showed elevated resistance to norfloxacin, ethidium bromide, acriflavine, and rhodamine 6G, from Pseudomonas aeruginosa YM64, a strain that lacks four major multidrug efflux pumps. The genes responsible for the resistance were mexHI-opmD. Elevated ethidium extrusion was observed with cells of YM644 and YM64 harboring a plasmid carrying the genes. Disruption of the genes in the chromosomal DNA of YM644 made the cells sensitive to the drugs.
Several mechanisms by which microorganisms overcome the toxicity of antimicrobial agents are known. Extrusion of toxic agents from microbial cells is one of these mechanisms. So far, five major groups of drug extrusion systems are known (2, 3, 11, 12, 15, 19): the MFS (major facilitator super), SMR (small multidrug resistance), RND (resistance nodulation cell division), MATE (multidrug and toxic compounds extrusion), and ABC (ATP binding cassette) families.
Pseudomonas aeruginosa shows significant degrees of intrinsic resistance to a wide variety of antimicrobial agents, including most β-lactams, fluoroquinolones, tetracycline, chloramphenicol, and erythromycin. At least five RND family drug efflux pumps are known to exist in cells of P. aeruginosa, MexAB-OprM (13), MexCD-OprJ (14), MexEF-OprN (6), MexXY-OprM (8), and MexJK-OprM (4). Very recently, it was reported that MexHI-OpmD is involved in resistance to vanadium (1). However, it is not known whether MexHI-OpmD functions as a multidrug efflux pump. Whole-genome sequence data of P. aeruginosa are now available (17) and suggest the presence of about 10 RND family multidrug efflux pumps in this microorganism. Previously, we reported the functional cloning of multidrug resistance genes in a drug-hypersensitive Escherichia coli mutant (8) and obtained many candidate plasmids conferring multidrug resistance. We cloned mexAB, mexCD, and mexXY, but not mexEF and others (8). The first three pumps are functional in wild-type P. aeruginosa (7, 9, 10, 11). It is likely that other genes for the RND family of multidrug efflux pumps are silent in wild-type P. aeruginosa.
It is very likely that mutation in the hitherto uncharacterized pumps leads to other multidrug resistance. Thus, the aim of our study was to analyze such uncharacterized multidrug pumps in P. aeruginosa. For this purpose, it would be valuable to isolate multidrug-resistant mutants from P. aeruginosa YM64, which lacks four major multidrug efflux operons (11). It is expected that silent genes for multidrug efflux pumps are functional in such mutants.
Isolation of a drug-resistant mutant.
Cells of P. aeruginosa strain YM64 (ΔmexAB-oprM ΔmexCD-oprJ ΔmexEF-oprN ΔmexXY) (11) were grown in Luria-Bertani (LB) medium (1% Bacto tryptone, 0.5% Bacto yeast extract, 1% NaCl [pH 7]) and were spread onto LB agar plates containing eightfold-higher concentrations than the MIC of norfloxacin. We obtained many mutants and selected the mutant YM644, which showed resistance to norfloxacin, ethidium bromide, acriflavine, and rhodamine 6G (Table 1). Thus, it seems that one of the uncharacterized multidrug efflux pumps, the gene or genes for which are silent, became functional.
TABLE 1.
MICs of various antimicrobial agents in strains of P. aeruginosa
| Drug | MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|
| PAO1 | YM64 | YM644 | YM64/pUPA2 | YM64/pUPA22 | PMX52 | PMX54 | |
| Norfloxacin | 1 | 0.06 | 1 | 1 | 0.25 | 0.06 | 0.06 |
| Ethidium bromide | 1,024 | 64 | 1,024 | 1,024 | 512 | 64 | 128 |
| Acriflavine | 4 | 512 | 512 | 16 | 4 | 8 | |
| Rhodamine 6G | 32 | 512 | 128 | 64 | 32 | 32 | |
| TPPCl | 64 | 512 | 256 | ||||
| Carbenicillin | 32 | 1 | 1 | 1 | 1 | ||
| Erythromycin | 64 | 8 | 8 | 8 | 8 | ||
| Gentamicin | 8 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Tetracycline | 16 | 0.25 | 0.25 | ||||
| Triclosan | 32 | 32 | 32 | 32 | 32 | 32 | 32 |
Cloning of genes responsible for the observed multidrug resistance.
As a second step, we cloned genes responsible for the multidrug resistance from the chromosomal DNA of the mutant YM644. By using the shotgun method with YM64 (11) as a host and pUCP20T (16, 20) as a cloning vector, we obtained many candidate hybrid plasmids. Cells of YM64 harboring a plasmid showed elevated resistance against several antimicrobial agents. We selected a plasmid, pUPA2, which carried a DNA insert derived from the chromosomal DNA of the mutant that was the shortest among the candidate plasmids tested. Cells of YM64/pUPA2 showed elevated resistance against norfloxacin, ethidium bromide, acriflavine, and rhodamine 6G (Table 1). The MICs of these drugs for cells of YM64/pUPA2 were almost same as those for cells of YM644. The levels of drug resistance to other antimicrobial agents tested (such as carbenicillin, gentamicin, and triclosan) were the same as those in YM64 and YM644. These results support the view that the cloned genes are responsible for the elevated drug resistance observed with YM644.
As a third step, we determined the nucleotide sequences of several portions of the pUPA2 plasmid. The results revealed that pUPA2 carries open reading frames PA4205 to PA4208, which have been reported in the whole-genome sequence (17). We constructed a plasmid, pUPA22, carrying open reading frames PA4206 (mexH), PA4207 (mexI), and PA4208 (opmD) (1), which was preceded by the promoter of the lactose operon of E. coli. Cells of P. aeruginosa YM64/pUPA22 showed elevated resistance against norfloxacin, ethidium bromide, acriflavine, and rhodamine 6G compared with YM64 (Table 1). Thus, it is clear that the mexHI-opmD genes are enough to confer the drug resistance. However, the MICs of these drugs observed with YM64/pUPA22 were lower than those of YM64/pUPA2.
Drug efflux.
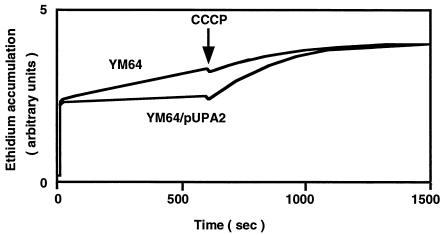
As a fourth step, we investigated drug efflux. Cells of YM64 showed a higher level of intracellular ethidium, suggesting no or very low efflux of ethidium from cells (Fig. 1). On the other hand, the intracellular ethidium level in YM64/pUPA2 was lower than that in YM64, suggesting efflux of ethidium. The intracellular ethidium level increased when an H+ conductor, carbonyl cyanide m-chlorophenylhydrazone (CCCP), was added to the assay mixture. Thus, energy-dependent efflux of ethidium occurred in cells of YM64/pUPA2. Addition of CCCP to a cell suspension of YM64 had little effect on the intracellular ethidium level, suggesting that energy-dependent efflux of ethidium is very low. We observed a similar intracellular ethidium level with YM644 compared to that with YM64/pUPA2 (data not shown). Thus, the energy-dependent efflux of ethidium is high in these two types of cells. These results support the idea that genes on the pUPA2 plasmid are coding for a drug efflux pump system.
FIG. 1.
Accumulation of ethidium in cells. Cells were grown in LB medium at 37°C and harvested at the middle of the exponential phase of growth. Cells were washed twice with Tanaka buffer (18) and suspended in the same buffer. Ethidium accumulation in cells was monitored continuously by measuring the fluorescence of ethidium at excitation and emission wavelengths of 500 and 580 nm, respectively. Ethidium bromide (final concentration, 100 μM) was added to the assay mixture to initiate the assay, and CCCP (final concentration, 100 μM) was added at time point indicated by a downward arrow.
mexHI-opmD deletion mutants.
The mexHI-opmD genes were removed from the chromosomal DNA of YM64 and YM644 by an Flp-FRT recombination system, as previously reported (5, 11), and PMX52 derived from YM64 and PMX54 derived from YM644 were obtained. Deletion of mexHI-opmD (BstI-BstI fragment was removed) was confirmed by Southern blot analysis with both the PMX52 and PMX54 strains (data not shown).
We measured the MICs of several antimicrobial agents with the deletion mutants and their parents (Table 1). Exactly the same MICs of norfloxacin, ethidium bromide, acriflavine, and rhodamine 6G were observed for YM64 and PMX52 (Table 1). This indicates that the MexHI-OpmD pump is not expressed or is not functional in YM64 cells. However, a great reduction in the MICs of norfloxacin, ethidium bromide, acriflavine, and rhodamine 6G was observed with PMX54 compared with its parent, YM644 (Table 1). Thus, we conclude that the mexHI-opmD element is expressed and the MexHI-OpmD pump is functional in cells of YM644. The MICs of ethidium bromide and acriflavine in PMX54, a mexHI-opmD-deleted derivative of YM644, were twofold higher than those in PMX52, a mexHI-opmD-deleted derivative of YM64. This twofold difference was reproducible. It should be pointed out that no differences in the MICs of carbenicillin, gentamicin, and triclosan were observed between the four strains (Table 1), as expected from the results shown in Table 1.
Susceptibility to vanadium and tetracycline.
Aendekerk et al. recently reported that MexHI-OpmD is responsible for resistance to vanadium (1). We investigated whether the MexHI-OpmD pump is involved in the vanadium resistance. We observed no detectable difference in vanadium susceptibility between YM64, YM644, PMX52, and PMX54 at concentrations of VOSO4 ranging from 1 to 4 mM (data not shown). Aendekerk et al. also reported changes in tetracycline susceptibility with a mutant with mutation of the MexHI-OpmD region (1); our data indicate that the MexHI-OpmD region is not involved in tetracycline susceptibility (Table 1). These discrepancies may be due to the difference in the strains used.
Acknowledgments
We thank M. Varela of Eastern New Mexico University for critical reading of the manuscript.
This research was supported by a grant from the Ministry of Education, Science, Sport and Culture of Japan.
REFERENCES
- 1.Aendekerk, S., B. Ghysels, P. Cornelis, and C. Baysse. 2002. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148:2371-2381. [DOI] [PubMed] [Google Scholar]
- 2.Bolhuis, H., H. W. van Veen, B. Poolman, A. J. Driessen, and W. N. Konings. 1997. Mechanisms of multidrug transporters. FEMS Microbiol. Rev. 21:55-84. [DOI] [PubMed] [Google Scholar]
- 3.Brown, M. H., I. T. Paulsen, and R. A. Skurray. 1999. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31:394-395. [DOI] [PubMed] [Google Scholar]
- 4.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 184:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 6.Köhler, T., M. Michea-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty, and J. C. Pechere. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23:345-354. [DOI] [PubMed] [Google Scholar]
- 7.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita, Y., N. Kimura, T. Mima, T. Mizushima, and T. Tsuchiya. 2001. Roles of MexXY- and MexAB-multidrug efflux pumps in intrinsic multidrug resistance of Pseudomonas aeruginosa PAO1. J. Gen. Appl. Microbiol. 47:27-32. [DOI] [PubMed] [Google Scholar]
- 10.Morita, Y., T. Murata, T. Mima, S. Shiota, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Induction of mexCD-oprJ operon for a multidrug efflux pump by disinfectants in wild-type Pseudomonas aeruginosa PAO1. J. Antimicrob. Chemother. 51:991-994. [DOI] [PubMed] [Google Scholar]
- 11.Morita, Y., Y. Komori, T. Mima, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2001. Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol. Lett. 202:139-143. [DOI] [PubMed] [Google Scholar]
- 12.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, and T. Yamasaki, S. Neshat, J. Yamagishi, X.-Z. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 15.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 17.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka, S., S. A. Lerner, and E. C. C. Lin. 1967. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J. Bacteriol. 93:642-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Veen, H. W., and W. N. Konings. 1997. Drug efflux proteins in multidrug resistant bacteria. Biol. Chem. 378:769-777. [PubMed] [Google Scholar]
- 20.West, S. E. H., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]