Abstract
The two genetic groups (oxy-1 and oxy-2) previously identified in the Klebsiella oxytoca taxon are recognizable by four independent molecular markers: (i) ERIC-1R profiles, (ii) 16S ribosomal DNA (rDNA) signature sequences, (iii) singular nucleotides in a defined fragment of the rpoB gene, and (iv) the type of the strain's blaOXY gene (i.e., blaOXY-1 or blaOXY-2). K. oxytoca strains SG266 and SG271 could not be classified into these genetic groups based on their ERIC-1R profile and blaOXY gene sequence. With regard to the gene identity percentages between the blaOXY-1 and blaOXY-2 gene groups (86.8% ± 0.4%) and within a blaOXY gene group (>99%), it was concluded that the blaOXY gene of strain SG271 was representative of a new blaOXY gene group (blaOXY-3), since the mean identity percentages between it and the two blaOXY gene groups were 85.5% ± 0.2% and 84.4% ± 0.4%, respectively. Since the corresponding percentages were 95.0% ± 0.4% and 86.2% ± 0.3% for strain SG266, it was impossible to classify its blaOXY gene, which was therefore named blaOXY-4. The 16S rDNA signature sequences of the two strains could be determined only after cloning experiments. The SG266 clones displayed the same signature sequence as that of the genetic group oxy-1, whereas the SG271 clones displayed three different 16S rDNA signature sequences that also differed from those of the two genetic groups. Singular nucleotides were found within the rpoB sequence of the two strains, allowing for their distinction from the two genetic groups. All of these results, combined with those previously obtained by the ERIC-1R PCR method, indicate that strain SG271 is representative of a new K. oxytoca genetic group (oxy-3), whereas strain SG266 could not be classified.
In 1974, Jain et al. (15) were able to distinguish the Oxytocum group from the Klebsiella genus by DNA-DNA hybrization. In spite of this discovery and also the fact that K. oxytoca strain 497-2 (ATCC 13182T) included in their analysis was a typical representative of this new taxon, these authors did not suggest a new genus of Enterobacteriaceae, concluding that such a suggestion might contribute to further confusion for bacteriologists, since Oxytocum organisms look like true Klebsiella with the exception of an indole reaction. This absence of a specific name is undeniably linked to the fact that true Klebsiella oxytoca strains had received multiple names (Klebsiella aerogenes and Klebsiella pneumoniae) until 1986, when Klesiella nomenclature was changed (2, 12). Although less common than K. pneumoniae in human infectious diseases (23), K. oxytoca was the object of particular interest since some strains were found as early as 1980 with a singular β-lactam resistance phenotype in comparison with K. pneumoniae strains. In fact, these K. oxytoca strains were found to be resistant to aztreonam in the first assays performed with this molecule by the manufacturer (24). Fifteen years later, Fournier et al. (9) demonstrated that this aztreonam resistance was due to mutations located in the promoter region of the gene (blaOXY) encoding the class A chromosomal β-lactamase (KOXY) of the resistant strains. Fournier et al. (11) also showed that the blaOXY genes were made up of two groups (blaOXY-1 and blaOXY-2) on the basis of the gene nucleotide sequence, with an identity percentage between the two gene groups of 87%. The corresponding KOXY β-lactamases were subsequently classified into the β-lactamase groups OXY-1 and OXY-2. More recently, an Enterobacteriaceae taxonomic analysis based on the sequence of certain fragments of the 16S ribosomal DNA (rDNA) and rpoB genes showed that K. oxytoca constitutes a monophylogenetic species clearly separated from K. pneumoniae (7). However, through different molecular studies, Brisse et al. (3) and Granier et al. (14) have shown that the K. oxytoca taxon is divided into two clades. We also determined that each clade corresponds to a K. oxytoca genetic group (oxy-1 or oxy-2) recognizable by four independent molecular markers: (i) characteristic bands in the profiles generated by the ERIC-1R PCR method, (ii) a signature sequence in a 16S rDNA fragment located from position 435 to position 458 according to the numbering of the Escherichia coli 16S rDNA sequence, (iii) singular nucleotides dispersed over the entire length of the 512-bp fragment of the rpoB gene shown by Mollet et al. (18) to identify enterobacterial species, and (iv) the type of the strain's blaOXY (blaOXY-1 or blaOXY-2) gene (14).
Using the ERIC-1R PCR method to classify a panel of K. oxytoca clinical isolates into these two genetic groups, we detected two particular K. oxytoca strains: SG266 and SG271. In fact, neither their ERIC-1R profiles nor the sequences of their blaOXY genes allowed us to classify them into the two K. oxytoca genetic groups previously described (13). Thus, the present study was carried out to determine whether these two isolates might be representative of two new K. oxytoca genetic groups.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains SG266 and SG271, identified by the routine method (API System; bioMérieux, Marcy l'Étoile, France) as K. oxytoca were obtained, both in 2000, from two patients hospitalized in the Saint Joseph Hospital in Paris (13). Strain SG266 infected a diabetic foot wound, and strain SG271 was isolated from peritoneal fluid. E. coli strain Epicurian Coli XL10-Gold and plasmid pPCR-Script (CamR; Stratagene, La Jolla, Calif.) were used for cloning experiments.
K. oxytoca strains ATCC 13182T and SC 10,436, which both harbored a blaOXY-2 gene controlled by a low and a strong promoter, respectively (12, 14), were used as control strains in β-lactam MIC tests.
Biochemical identification.
Strains SG266 and SG271 were resubmitted to biochemical identification by using the BIOTYPE-100 carbon source strip system (bioMérieux). This test was performed by the French Enterobacteriaceae Reference Center (Institut Pasteur, Paris, France).
β-Lactam susceptibility.
β-Lactam susceptibility was determined by the agar dilution method on Mueller-Hinton agar with a Steers multiple inoculator and 104 CFU per spot. The following antibiotics were tested: amoxicillin (GlaxoSmithKline, Marly-le-Roi, France); ticarcillin (GlaxoSmithKline), alone or combined with 2 μg of clavulanate (GlaxoSmithKline)/ml; piperacillin (Wyeth-Lederle, Paris-La Defense, France), alone or combined with 4 μg of tazobactam (Wyeth-Lederle)/ml; cephalothin (Lilly, Suresnes, France); cefotaxime (Aventis, Paris, France); ceftazidime (GlaxoSmithKline); cefepime (Bristol-Myers Squibb, Paris-La Defense, France); aztreoman (Sanofi Winthrop, Gentilly, France); cefoxitine (Panpharma, Fougeres, France); and imipenem (Merck Sharp & Dohme-Chibret, Paris, France). The results were interpreted according to the French Antibiogram Committee recommendations (1).
blaOXY gene and KOXY β-lactamase sequence comparison.
The blaOXY genes of strains SG266 and SG271 (GenBank accession numbers AY077481 and AF491278, respectively) and the corresponding KOXY β-lactamases were aligned by using Align software (20), with the blaOXY genes and the corresponding proteins registered under the following GenBank accession numbers: Z30177, Y17715, AY077486, AY077484, AY077483, AY077482, and M27459 for the blaOXY-1 genes and Z49084, AF473577, AY055205, AY077489, AY077488, AY077487, AY077485, D84548, and Y17714 for the blaOXY-2 genes. Three blaOXY-1 genes (strains 756, 563, and 1879-77) and three blaOXY-2 genes (strains K1794, 11V, and KER) that had been published (10, 22) but not submitted to GenBank were also included in the comparative analysis.
Preparation of crude extracts and isoelectrofocusing.
This experiment was performed as previously described (17).
The pI values of the β-lactamases produced by strains SG266 and SG271 were determined by comparison with the pI values of known β-lactamases—OXY-2-1 (pI 5.2), TEM-1 (pI 5.4), TEM-2 (pI 5.6), PSE-2 (pI 5.7), SHV-3 (pI 7), OXY-1-1 (pI 7.5), ACC-1 (pI 7.75), and SHV-5 (pI 8.2)—and isoelectrofocusing standards (Bio-Rad Laboratories, Hercules, Calif.).
KOXY β-lactamase phylogenetic tree.
The amino acid sequences of the KOXY β-lactamases previously published (2, 9-12, 14, 16, 22, 25) and those of the β-lactamases of strains SG266 and SG271 were aligned by using Dialign 2 software (19). The KOXY β-lactamase phylogenetic tree was built with TreeTop software from GeneBee services (4).
PCR, sequencing, and comparison of rpoB and 16S rDNA genes.
The 512-bp fragment of the rpoB gene and the 387-bp fragment of the 16S rDNA gene, allowing for the identification of enterobacterial species (7) and K. oxytoca genetic groups, were amplified from strains SG266 and SG271 and sequenced as previously described (14). The rpoB sequences were then directly compared by using Align software (20) to the previously published rpoB consensus sequences characteristic of the K. oxytoca genetic groups oxy-1 and oxy-2 (14). For the 16S rDNA fragment, the sequences were compared to the 16S rDNA consensus sequences only after cloning experiments.
16S rDNA cloning experiments and analysis of recombinant plasmids.
The 16S rDNA amplified product obtained from strains SG266 and SG271was purified and then cloned into pPCR-Script cloning vector according to the manufacturer's recommendations (Stratagene, La Jolla, Calif.). The recombinant plasmids were transferred into E. coli XL10-Gold by using the heat shock method. Transformants were selected onto agar plates containing chloramphenicol (30 mg/liter), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 mg/liter), and IPTG (isopropyl-β-d-thiogalactopyranoside; 40 mg/liter), and those displaying a white color were analyzed. Thus, the fragment inserted into the recombinant plasmids was amplified by using the plasmidic T3 and T7 primers; fragments with the expected size were then sequenced.
GenBank accession numbers.
The rpoB gene partial sequences of strains SG266 and SG271 have been registered in GenBank database under accession numbers AF491282 and AF491280, respectively.
RESULTS
blaOXY gene and KOXY β-lactamase comparison.
As indicated in Table 1, pairwise comparative analysis of the blaOXY gene of strains SG266 and SG271 and the blaOXY genes previously published, i.e., 10 blaOXY-1 and 12 blaOXY-2 genes showed that the mean percentages of blaOXY gene identity were 95.0% ± 0.4% and 86.2% ± 0.3% for strain SG266 and blaOXY-1 and blaOXY-2 genes, respectively, and 85.5% ± 0.2% and 84.4% ± 0.4%, respectively, for strain SG271. These percentages were 86.8% ± 0.4% between the blaOXY-1 and the blaOXY-2 genes and only 85.8% between the blaOXY genes of strains SG266 and SG271. The comparison of the KOXY β-lactamases encoded by all of these blaOXY genes allowed us to determine that certain amino acids (n = 34) located at positions not previously defined as being involved in the enzymatic active site (8) were singular for one of the four β-lactamase types: OXY-1 group, OXY-2 group, the KOXY β-lactamase of strain SG266, and the KOXY β-lactamase of strain SG271 (Table 2) A particular amino acid was more often observed in the KOXY β-lactamase of strain SG271 (n = 15) and the OXY-2 group (n = 14) than in the OXY-1 group (n = 3 or 5) or the KOXY β-lactamase of strain SG266 (n = 3). As indicated in Table 2, the pI values of the β-lactamases of strains SG266 and SG271 were 7.7 and 6.7, respectively.
TABLE 1.
Identity percentages between the nucleotide sequences of the blaOXY and rpoB genes of K. oxytoca strains SG266 and SG271 and the previously published blaOXY genes and the rpoB consensus sequences characteristic of the K. oxytoca genetic groups oxy-1 and oxy-2
| Genetic group or strain (n) | % Identity ± SD
|
|||||
|---|---|---|---|---|---|---|
| Genetic group oxy-2
|
Strain SG266
|
Strain SG271
|
||||
| blaOXY-2 | rpoB consensus | blaOXY | rpoB | blaOXY | rpoB | |
| Genetic group oxy-1 | ||||||
| blaOXY-1 (10) | 86.8 ± 0.4 | 95.0 ± 0.4 | 85.5 ± 0.2 | |||
| rpoB consensus | 96.7 | 96.5 | 96.5 | |||
| Genetic group oxy-2 | ||||||
| blaOXY-2 (12) | 86.2 ± 0.3 | 84.4 ± 0.4 | ||||
| rpoB consensus | 95.3 | 97.3 | ||||
| Strain SG266 | ||||||
| blaOXY | 85.8 | |||||
| rpoB | 95.9 | |||||
TABLE 2.
pI values and discriminant amino acids for OXY-1 and OXY-2 β-lactamase groups and the β-lactamases of K. oxytoca strains SG266 and SG271
| β-Lactamase group or strain (n) | pI value(s) | Amino acida at position (Ambler numbering):
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 (−) | 9 (−) | 11 (−) | 12 (8) | 15 (11) | 17 (13) | 22 (18) | 25 (21) | 29 (25) | 30 (26) | 31 (27) | 32 (28) | 33 (29) | ||
| OXY-1 group (8)b | 7.1, 7.5, 8.2, 8.8 | S | T | L | M | A | V | A | S | S | A/V | D | A | I |
| OXY-2 group (9)c | 5.2, 5.7, 6.4, 6.5, 6.8 | S | I | M | L | del | V | A | A | S | T | D | A | I |
| SG266 | 7.7 | S | S | L | M | del | V | A | S | S | A | D | T | L |
| SG271 | 6.7 | T | S | L | I | del | L | C | S | N | A | del | del | I |
| Amino acida at position (Ambler numbering):
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 38 (34) | 43 (39) | 44 (40) | 54 (50) | 56 (52) | 75 (72) | 90 (−) | 101 (98) | 103 (100) | 111 (108) | 129 (128) | 140 (137) | 143 (140) | 161 (158) | 168 (165) | 171 (168) | 134 (181) | 194 (191) | 230 (227) | 270 (269) | 278 (277) |
| A | R | S | N | A | G | N | K | S | I | A | K | S | V | T | A | T | R | A | Q | E/K |
| T | R | S | N | A | S | N | N | A | I | T | L | G | A | T | T | S | R | E | Q | E |
| A | R | S | N | A | G | H | K | S | I | A | K | G | V | M | A | T | R | A | Q | E |
| A | S | T | D | T | G | N | K | S | V | A | K | G | V | T | A | T | H | A | L | D |
Amino acid numbering is based on the sequence of the OXY-1 β-lactamases.
Strains SL781 (10), KH66 (25), SG344 (14), 756 (10), SG49 (14), 563 (10), 1879-77 (10), and SG337 (14).
Strains SC 10,436 (12), KoKER (22), 11V (10), D488 (10), SL911 (10), ATCC 13182 (14), SG4 (14), SG77 (14), and SG176 (14). Amino acids in boldface are specific for genetic groups or strains.
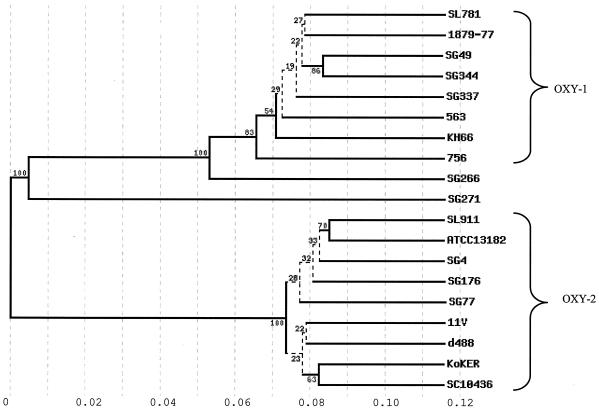
According to the phylogenetic tree constructed from the amino acid sequence of all of these KOXY β-lactamases, we observed three phylogenetically distant branches (Fig. 1). The first branch included all of the OXY-1 β-lactamases tested and the KOXY β-lactamase of strain SG266 which was, however, the most distant β-lactamase in this β-lactamase group. The second branch included only the KOXY β-lactamase of strain SG271, and the third branch included exclusively the OXY-2 β-lactamases tested. This subdivision was obtained with bootstrap values of 100%.
FIG. 1.
KOXY β-lactamase phylogram. The tree was built from the KOXY β-lactamase sequences of 19 strains (as indicated on the right side of the figure). Eight β-lactamases belong to the OXY-1 group, and nine belong to the OXY-2 group. Numbers within the phylogram indicate the bootstrap values of branching. The distance ladder is indicated under the tree.
β-Lactam MICs.
As indicated in Table 3 K. oxytoca strains SG266 and SG271 were resistant to amoxicillin and ticarcillin but susceptible to these two molecules when they were combined with 2 μg of clavulanate/ml and were also susceptible also to all other β-lactam molecules tested.
TABLE 3.
β-Lactam MICs for K. oxytoca strains ATCC 13182T, SC 10,436, SG266, and SG271
| β-Lactam | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| ATCC 13182Ta | SC 10,436b | SG266 | SG271 | |
| Amoxicillin | 128 | >1,024 | 64 | 16 |
| + 2 μg of clavulanate/ml | 1 | 128 | 0.5 | 0.25 |
| Ticarcillin | 32 | >1,024 | 64 | 16 |
| + 2 μg of clavulanate/ml | 1 | 256 | 2 | 0.5 |
| Piperacillin | 2 | 512 | 2 | 1 |
| + 4 μg of tazobactam/ml | 0.5 | 256 | 0.5 | 0.12 |
| Cephalothin | 2 | 64 | 2 | 2 |
| Cefoxitin | 0.5 | 1 | 1 | 0.5 |
| Cefotaxime | ≤0.06 | 2 | ≤0.06 | ≤0.06 |
| Ceftazidime | 0.12 | 1 | ≤0.06 | ≤0.06 |
| Cefepime | ≤0.06 | 2 | ≤0.06 | ≤0.06 |
| Aztreonam | ≤0.06 | 32 | ≤0.06 | ≤0.06 |
| Imipenem | 0.5 | 0.5 | 0.5 | 0.5 |
Strain ATCC 13182T (14) harboring a blaOXY-2 gene under the control of a wild type promoter.
Strain SC 10,436 (12) harboring a blaOXY-2 gene under the control of a mutated promoter responsible for the β-lactamase overproduction.
rpoB and 16S rDNA sequence comparison.
The comparison (Table 1) of the 512-bp fragment of the rpoB gene of strains SG266 and SG271 with the two rpoB consensus sequences previously defined for the strains belonging to the K. oxytoca genetic groups oxy-1 and oxy-2, respectively, showed that the percentage of identity was higher between strain SG266 and the oxy-1 genetic group (96.5%) than between strain SG266 and the oxy-2 genetic group (95.3%) and, inversely, higher between strain SG271 and the oxy-2 genetic group (97.3%) than between strain SG271 and the oxy-1 genetic group (96.5%). The percentage of the rpoB gene identity between strains SG266 and SG271 was only 95.9%, whereas this percentage was 96.7% between the two rpoB consensus sequences (Table 1). The nucleotide differences observed all along the 512-bp rpoB fragment between strain SG266, strain SG271, the consensus sequence of the genetic group oxy-1, and that of the genetic group oxy-2 resulted in several singular nucleotides at given positions according to the two genetic groups and the two strains (Table 4) Thus, C162, T258, C264, C270, C276, and G285 were only observed in the rpoB consensus sequence of the genetic group oxy-1, whereas A3, C114, C123, and T339 were only observed in that of the genetic group oxy-2. Six (T2, T3, A36, G84, A108, and G141) and nine (C87, T108, T168, C192, T255, G270, C372, T468, and T489) other particular amino acids were observed in the 512-bp rpoB fragments of strains SG266 and SG271, respectively.
TABLE 4.
Nucleotide variations within the 512-bp fragment of the rpoB gene of K. oxytoca belonging to the oxy-1 and oxy-2 genetic groups and of strains SG271 and SG266
| Genetic group or strain | Nucleotide at positiona:
|
||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 6 | 24 | 36 | 84 | 87 | 96 | 108 | 114 | 120 | 123 | 141 | 162 | 168 | 192 | 225 | 246 | 255 | 258 | 264 | 270 | 276 | 285 | 288 | 294 | 300 | 339 | 351 | 372 | 447 | 468 | 486 | 489 | 504 | |
| oxy-1 | C | G | A | C | G | A | T | C/T | G | T | C | T | A | C | C | G | C/T | G | C | T | C | C | C | G | T | T | A | C | T | G | C | A | A | C | G/A |
| oxy-2 | C | A | G/A | C | G | A | T | T | G | C | T | C | A | T | C | G | C | G | C | C | T | T | T | T | C | C | G | T | T | G | C/T | A | T | C | G |
| SG266 | T | T | A | T | A | G | T | C | A | T | T | T | G | T | C | G | C | T | C | C | T | T | T | T | C | C | G | C | C | G | C | A | A | C | G |
| SG271 | C | G | A | T | G | A | C | C | T | T | C | T | A | T | T | C | C | T | T | C | T | G | T | T | T | T | A | C | C | C | T | T | T | T | G |
Numbering starts at codon 500 of the RpoB protein of E. coli (18). Nucleotides in boldface are characteristic of genetic groups or strains. A shill indicates the variability of the sequence within a genetic group, with the first nucleotide being the more frequent (consensus sequence).
Concerning the sequence of the 387-bp 16S rDNA fragments of strains SG266 and SG271, we observed (Table 5) different nucleotide possibilities on the chromatograph, exactly at the nucleotide positions that allowed us to distinguish (signature sequences) the two 16S rDNA consensus sequences characteristic of the two K. oxytoca genetic groups (14). After we cloned the 387-bp fragment, which was amplified from strains SG266 and SG271, we analyzed the sequence of the 387-bp fragment from five clones derived from strain SG266 and four clones derived from strain SG271. The five clones derived from strain SG266 displayed an identical signature sequence corresponding to that previously defined for the K. oxytoca genetic group oxy-1 (Table 5), whereas the four clones derived from strain SG271 displayed three different signature sequences that were, moreover, different from those previously defined for the two genetic groups of K. oxytoca (Table 5). Pairwise analysis of the sequences of the 16S rDNA fragment of the three different clones obtained from strain SG271 and the16S rDNA consensus sequence of each genetic group showed (Table 5) that clone C had a high percentage of identity with the 16S rDNA consensus sequence of the genetic group oxy-1, whereas clone D displayed a similarly high percentage of identity with the 16S rDNA consensus sequence of the genetic group oxy-2. Clones A and B, which showed the same 16S rDNA signature sequence, displayed a lower identity percentages with each 16S rDNA consensus sequence, a percentage that was similar to that observed between the two 16S rDNA consensus sequences (Table 5).
TABLE 5.
16S rDNA signature sequences and percentages of the 16S rDNA fragment homology between K. oxytoca genetic groups, strains SG266 and SG271, and their 16S rDNA clones
| Genetic group or strain | 16S rDNA signature sequence between positions 435 and 458a | % Identity between the 16S rDNA fragment and:
|
||||
|---|---|---|---|---|---|---|
| oxy-2 | SG266 clone A | SG271 clone A | SG271 clone C | SG271 clone D | ||
| oxy-1 | CGATA-(14n)-GTCG | 98.1 | 100 | 98.6 | 99.5 | 98.4 |
| oxy-2 | GAGTG-(14n)-ATTC | 98.1 | 98.9 | 97.8 | 99.7 | |
| SG266 | G/C A/G A/G T A-(14n)-G/A T T/C C/G | |||||
| SG266 clone Ab | CGATA-(14n)-GTCG | 98.6 | 99.5 | 98.4 | ||
| SG271 | C/GG/A A/T/G T A-(14n)-A/G T/G T/C C/G | |||||
| SG271 clone Ac | GGATA-(14n)-ATTC | 98.1 | 99.2 | |||
| SG271 clone C | CGTTA-(14n)-GGCG | 98.1 | ||||
| SG271 clone D | GAGTA-(14n)-ATTC | |||||
According to the numbering of the E. coli 16S rDNA sequence (5).
Clones B, C, D, and E from strain SG266 displayed the same sequence as clone A.
Clone B from strain SG271 displayed the same sequence as clone A.
Extensive biochemical analysis of strains SG266 and SG271.
Although all of the genetic analyses performed indicated that strains SG266 and SG271 belong to the K. oxytoca taxon, we decided to submit them to an extensive biochemical analysis since they displayed particular features regarding the genes studied.
Although strain SG266 displayed negative tests for three biochemical reactions normally positive for K. oxytoca strains (meso-tartrate, trans-aconitate, and dl-glycerate), it was confirmed as belonging to the K. oxytoca taxon. Strain SG271, which displayed numerous negative reactions (notably for meso-tartrate, d-malate, trans-aconitate, tricarballylate, putrescine, succinate, fumarate, dl-glycerate, d-glucosamine, and l-aspartate), could only be identified as Klebsiella but without species definition.
DISCUSSION
We have previously determined that the K. oxytoca taxon is divided into two genetic groups (oxy-1 or oxy-2) recognizable by four independent molecular markers (14) and have proved that the genetic group classification made on the basis of the analysis of one of these four markers was confirmed by the secondary analysis of another marker. Thus, we have classified K. oxytoca strain ATCC 13182T into the oxy-2 genetic group on the basis of its 16S rDNA and rpoB sequences available in GenBank and then confirmed this classification by sequencing its blaOXY gene (14). We have also classified 15 clinical isolates identified as K. oxytoca through routine biochemical analysis into either of the two genetic groups on the basis of their ERIC-1R profile and then confirmed this classification by sequencing their blaOXY genes (13). However, we were unable to classify strains SG266 and SG271 into the two K. oxytoca genetic groups on the basis of their ERIC-1R profiles and the sequences of their blaOXY genes (13). Subsequently, the present study was carried out to determine whether these two strains could be representative of two new K. oxytoca genetic groups.
Taking into consideration the fact that the blaOXY gene of strain SG271 displayed a lower identity percentage with the blaOXY-1 gene group (85.5% ± 0.2%) and with the blaOXY-2 gene group (84.4% ± 0.4%) than that observed between these two blaOXY gene groups (86.8% ± 0.4%), we can reasonably conclude that the blaOXY gene of strain SG271 is representative of a new blaOXY gene group that we therefore suggest naming blaOXY-3. This lower gene identity percentage resulted in the creation of a new branch of the KOXY β-lactamase phylogenetic tree, to which only the chromosomal β-lactamase of strain SG271 was attached. Thus, the OXY-3 β-lactamase of strain SG271 further enhances the aspect of the phylogenetic tree of KOXY β-lactamases. Regarding the sequence of the 512-bp fragment of the rpoB gene of strain SG271, we found six positions for which there were nucleotides specific for strain SG271 allowing us to distinguish this strain from the two genetic groups previously described (13). The third marker previously defined as being characteristic of the K. oxytoca genetic groups, namely, the 16S rDNA signature sequence, displayed new genetic features in strain SG271. In fact, by cloning the 387-bp 16S rDNA fragment from strain SG271, we were able to find three different signature sequences out of the four sequenced clones. This finding suggests the presence of at least three 16S rRNA operons in K. oxytoca and confirmed the existence of variations within the 16S rRNA operons of a same strain, as previously described for a very few bacterial species (6, 21, 26). Moreover, we found that the three different signature sequences also differed from either of the two 16S rDNA signature sequences characteristic of the two K. oxytoca genetic groups. All of these findings, combined with the fact that the ERIC-1R profile of strain SG271 did not display the oxy-1 or oxy-2 genetic group-specific bands (13), led us to conclude that we have discovered in strain SG271 a strain representative of a third genetic group in the K. oxytoca taxon. We suggest that this third genetic group be called oxy-3. The last point that we can emphasize about strain SG271 is the fact that the biochemical analysis performed by using 100 molecules as carbon source was able to identify it at only the genus level, Klebsiella, whereas all of the molecular studies that we carried out allowed for a species identification.
With regard to β-lactam susceptibility, strain SG271 displayed a β-lactam resistance pattern identical to that displayed by K. oxytoca strains normally producing β-lactamase. This was also the case of strain SG266, which was confirmed to be K. oxytoca via extensive biochemical analysis. However, the results obtained for this strain with regard to the four molecular markers characteristic of the K. oxytoca genetic groups did not allow us to classify it. Indeed, even if its blaOXY gene displayed a high identity percentage with the genes of the blaOXY-1 group (95.0% ± 0.4%) and a low percentage with the genes of the blaOXY-2 group (86.2% ± 0.3%), this did not allow us to classify the SG266 strain's blaOXY gene in the blaOXY-1 group, since we had previously shown that the identity percentage within a blaOXY group was >99% (14). We therefore suggest that this gene be called blaOXY-4. Cloning experiments of the 387-bp 16S rDNA fragment of strain SG266 resulted in five clones displaying identical signature sequences that perfectly matched those of the strains belonging to the oxy-1 genetic group. Among the clones tested, we did not find any that were responsible for the variations observed in the sequence of the fragment amplified and directly sequenced from strain SG266. This result suggests that the sequences found in the five clones are probably dominant among the 16S rRNA operons of strain SG266. The two remaining oxy genetic group markers studied for strain SG266, ERIC-1R profile and singular nucleotides in the 512-bp fragment of the rpoB gene, not only confirmed that it was not possible to classify strain SG266 into the oxy-1 genetic group but also suggested that strain SG266 could belong to a subgenetic group. Indeed, on the basis of these two molecular markers, strain SG266 can clearly be distinguished from those belonging to either of the three oxy genetic groups that have been defined in the K. oxytoca taxon up to now. Finally, the possibility that strain SG266 might belong to a subgenetic group was also suggested by the position of its β-lactamase in the β-lactamase phylogenetic tree.
Interestingly, by discovering the two strains SG266 and SG271, we observed that the number of singular nucleotides, by which the two rpoB consensus sequences could have been differentiated (14), decreased. In fact, instead of the 17 nucleotides found to be specific for the recognition of the two genetic groups oxy-1 and oxy-2, only 6 remained specific for the oxy-1 genetic group and only 4 remained specific for the oxy-2 genetic group. However, six and nine became specific for strains SG266 and SG271, respectively. Such a discriminatory power cannot be attributed to the 16S rDNA signature sequence since the 16S rDNA fragment studied did not allow for the differentiation of strain SG266 from those of the oxy-1 genetic group.
Overall, the present study confirms that the ERIC-1R PCR method is a powerful tool to detect particular strains in the K. oxytoca taxon (13) and that the chromosomal bla gene of K. oxytoca, although it is not classified within the housekeeping gene family, is able, like housekeeping genes, to trace the bacterial diversification and evolution in the K. oxytoca taxon (14).
Acknowledgments
This study was supported by a grant from the French β-lactamase network (Ministère de la Recherche, Paris, France).
We thank Nadine Honoré for help with methodology choices.
REFERENCES
- 1.Anonymous. 2003. Comité de l'antibiogramme de la Societé Française de Microbiologie report 2003. Int. J. Antimicrob. Agents. 21:364-391. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa, Y., M. Ohta, N. Kido, M. Mori, H. Ito, T. Komatsu, Y. Fujii, and N. Kato. 1989. Chromosomal beta-lactamase of Klebsiella oxytoca, a new class A enzyme that hydrolyzes broad-spectrum beta-lactam antibiotics. Antimicrob. Agents Chemother. 33:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brisse, S., and J. Verhoef. 2001. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing, and automated ribotyping. Int. J. Syst. E vol. Microbiol. 51:914-925. [DOI] [PubMed] [Google Scholar]
- 4.Brodsky, L. I., A. V. Vasiliev, Y. L. Kalaidzidis, Y. S. Osipov, R. L. Tatuzov, and S. I. Feranchuk. 1992. GeneBee: the program package for biopolymer structure analysis. Dimacs 8:127-139. [Google Scholar]
- 5.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cilia, V., B. Lafay, and R. Christen. 1996. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol. Biol. E. 13:451-461. [DOI] [PubMed] [Google Scholar]
- 7.Drancourt, M., C. Bollet, A. Carta, and P. Rousselier. 2001. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov., and Raoultella planticola comb. nov. Int. J. Syst. Evol. Microbiol. 51:925-932. [DOI] [PubMed] [Google Scholar]
- 8.Farzaneh, S., J. Peduzzi, L. Sofer, A. Reynaud, M. Barthelemy, and R. Labia. 1997. Characterization and amino acid sequence of the OXY-2 group for beta-lactamase of pI 5.7 isolated from aztreonam-resistant Klebsiella oxytoca strain HB60. J. Antimicrob. Chemother. 40:789-795. [DOI] [PubMed] [Google Scholar]
- 9.Fournier, B., C. Y. Lu, P. H. Lagrange, R. Krishnamoorthy, and A. Philippon. 1995. Point mutation in the Pribnow box, the molecular basis of beta-lactamase overproduction in Klebsiella oxytoca. Antimicrob. Agents Chemother. 36:1365-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fournier, B., and P. H. Roy. 1997. Variability of chromosomally encoded beta-lactamases from Klebsiella oxytoca. Antimicrob. Agents Chemother. 41:1641-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier, B., P. H. Roy, P. H. Lagrange, and A. Philippon. 1996. Chromosomal beta-lactamase genes of Klebsiella oxytoca are divided into two main groups, blaOXY-1 and blaOXY-2. Antimicrob. Agents Chemother. 40:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granier, S. A., K. Bush, V. Leflon-Guibout, F. W. Goldstein, and M. H. Nicolas-Chanoine. 2002. The extended-spectrum K1 beta-lactamase from Klebsiella oxytoca SC 10,436 is a member of the blaOXY-2 family of chromosomal Klebsiella enzymes. Antimicrob. Agents Chemother. 46:2056-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granier, S. A., V. Leflon-Guibout, F. W. Goldstein, and M. H. Nicolas-Chanoine. 2003. Enterobacterial repetitive intergenic consensus 1R PCR Assay for detection of Raoultella sp. isolates among strains identified as K. oxytoca in clinical laboratory. J. Clin. Microbiol. 41:1740-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granier, S. A., L. Plaisance, V. Leflon-Guibout, E. Lagier, S. Morand, F. W. Goldstein, and M. H. Nicolas-Chanoine. 2003. Recognition of two genetic groups in Klebsiella oxytoca taxon on the basis of the chromosomal beta-lactamase and housekeeping gene sequences as well as ERIC PCR typing. Int. J. Syst. E vol. Microbiol. 53:661-668. [DOI] [PubMed] [Google Scholar]
- 15.Jain, K., K. Radsak, and W. Mannheim. 1974. Differentiation of the oxytocum group from Klebsiella by deoxyribonucleic acid-deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 24:402-407. [Google Scholar]
- 16.Kimura, K., Y. Arakawa, S. Ohsuka, H. Ito, K. Suzuki, H. Kurokawa, N. Kato, and M. Ohta. 1996. Molecular aspects of high-level resistance to sulbactam-cefoperazone in Klebsiella oxytoca clinical isolates. Antimicrob. Agents Chemother. 40:1988-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariotte, S., P. Nordmann, and M. H. Nicolas. 1994. Extended-spectrum beta-lactamase in Proteus mirabilis. J. Antimicrob. Chemother. 33:925-935. [DOI] [PubMed] [Google Scholar]
- 18.Mollet, C., M. Drancourt, and D. Raoult. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26:1005-1011. [DOI] [PubMed] [Google Scholar]
- 19.Morgenstern, B. 1999. Dialign 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15:203-210. [DOI] [PubMed] [Google Scholar]
- 20.Myers, E. W., and W. Miller. 1988. Optimal alignments in linear space. Comput. Appl. Biosci. 4:11-17. [DOI] [PubMed] [Google Scholar]
- 21.Reischl, U., K. Feldmann, L. Naumann, B. J. Gaugler, B. Ninet, and B. Hirschel. 1998. 16S rRNA sequence diversity in Mycobacterium celatum strains caused by the presence of two different copies of 16S rRNA gene. J. Clin. Microbiol. 36:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirot, D., R. Labia, P. Pouedras, C. Chanal-Claris, C. Cerceau, and J. Sirot. 1998. Inhibitor-resistant OXY-2 derived beta-lactamase produced by Klebsiella oxytoca. Antimicrob. Agents Chemother. 42:2184-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sirot, J., M. H. Nicolas-Chanoine, H. Chardon, J. L. Avril, C. Cattoen, J. C. Croix, H. Dabernat, T. Fosse, J. C. Ghnassia, E. Lecaillon, A. Marmonier, M. Roussel-Delvallez, C. J. Soussy, A. Trevoux, F. Vandenesh, C. Dib, N. Moniot-Ville, and Y. Rezvani. 2002. Susceptibility of Enterobacteriaceae to beta-lactam agents and fluoroquinolones: a 3-year survey in France. Clin. Microbiol. Infect. 8:207-213. [DOI] [PubMed] [Google Scholar]
- 24.Sykes, R. B., D. P. Bonner, K. Bush, and N. H. Georgopapadakou. 1982. Aztreonam (SQ 26,776), a synthetic monobactam specifically active against aerobic gram-negative bacteria. Antimicrob. Agents Chemother. 21:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, S. W., K. Dornbusch, and G. Kronvall. 1999. Genetic characterization of resistance to extend-spectrum beta-lactams in Klebsiella oxytoca isolates recovered from patients with septicemia at hospitals in the Stockholm area. Antimicrob. Agents Chemother. 43:1294-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]