Abstract
Klebsiella pneumoniae FFUL 22K was isolated in April 1999 from the urine of an intensive care unit patient in Portugal. The strain showed an extended-spectrum cephalosporin resistance profile. A typical synergistic effect between cefotaxime or cefepime and clavulanic acid was observed. An Escherichia coli transformant displayed a similar resistance phenotype and harbored a ca. 9.4-kb plasmid (p22K9). Cloning experiments revealed that the extended-spectrum β-lactamase was encoded by blaGES-1, previously described in class 1 integrons from K. pneumoniae ORI-1 and Pseudomonas aeruginosa Pa695. Further sequence analysis demonstrated that the blaGES-1 gene cassette was located on a new class 3 integron. The integron was 2,863 bp long and consisted of an intI3 integrase gene, an attI3 recombination site, two promoter regions, and two gene cassettes. The IntI3 integrase was 98.8% identical to that of Serratia marcescens AK9373. The blaGES-1 gene cassette was inserted at the attI3 site. The second gene cassette was the result of a fusion event between blaOXA-10-type and aac(6′)-Ib gene cassettes and conferred resistance to kanamycin. This is the second class 3 integron reported and the first time that the blaGES-1 gene cassette has been found on an integron belonging to this class, highlighting the considerable heterogeneity of their genetic environment and the spread of gene cassettes among different classes of integrons.
Many of the antibiotic resistance genes found in clinical isolates of gram-negative bacteria are contained in discrete mobile elements known as gene cassettes, located in integrons (19, 20, 21, 45). The integron encodes a site-specific recombinase (IntI) that belongs to a distinct family of the tyrosine recombinase superfamily (10), responsible for the insertion of gene cassettes at attI, and also provides the promoter responsible for expression of the cassette-encoded genes (5).
Ten classes of integrons have been identified, five of them associated with gene cassettes that codify antibiotic resistance (2, 7, 19, 21, 22, 23, 30, 37, 45; GenBank accession number AJ277063). However, the integrons most commonly isolated from resistant clinical isolates of members of the family Enterobacteriaceae belong to class 1. To date, only a single class 3 integron, isolated from a carbapenem-resistant Serratia marcescens strain, has been described (2). Characterization of this integron by Collis et al. (10) revealed that the integron module, consisting of the intI3 gene, attI3 site, and Pc promoter, is configured in the same way as the class 1 integron module.
Among the β-lactamase genes, blaVEB-1 was the first gene cassette identified encoding a class A enzyme that possesses extended-spectrum properties (34, 35, 44). Subsequently blaGES-1 and its homologous blaIBC-1, blaGES-2 and blaIBC-2 gene cassettes were found in several class 1 integrons (15, 16, 29, 40, 41, 42).
In this work, we report the analysis of the Klebsiella pneumoniae FFUL 22K clinical isolate exhibiting extended-spectrum cephalosporin resistance. Cloning experiments revealed the presence of a new class 3 integron on a small plasmid.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| K. pneumoniae FFUL 22K | Extended-spectrum cephalosporin-resistant clinical isolate | This study |
| E. coli DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Gibco-BRL |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| p22K9 | Natural plasmid from K. pneumoniae FFUL 22K | This study |
| pBK-CMV | Cloning vector, Kanr Neor | Stratagene |
| pMFA-1 | Recombinant pBK-CMV plasmid with a 1,284-bp HindIII insert from p22K9 containing part of the class 3 integron and the blaGES-1 gene | This study |
| pMFA-2 | Recombinant pBK-CMV plasmid with a 1,388-bp amplicon containing the entire Pc promoter region from the integron and the blaGES-1 gene | This study |
Antibiotic susceptibility testing.
The antibiotic susceptibilities of K. pneumoniae clinical isolate and Escherichia coli recombinant strains were determined by the disk diffusion method (36). The double-disk synergy test between clavulanic acid and extended-spectrum cephalosporins was used to detect production of extended-spectrum β-lactamases (4). The MICs of selected β-lactam antibiotics were determined by the E-test method (AB Biodisk, Solna, Sweden).
Isoelectric focusing.
Analytical isoelectric focusing of crude cell extracts from K. pneumoniae FFUL 22K and E. coli transformants was performed in polyacrylamide gels containing ampholytes (pH 5.0 to 8.0; Amersham Biosciences Europe, Lisbon, Portugal) with a Multiphor II apparatus (Amersham Biosciences Europe, Lisbon, Portugal). The focused β-lactamases were detected after overlaying the gels with nitrocefin (Calbiochem EMD Biosciences, San Diego, Calif.), and the isoelectric points were determined and compared to those of known β-lactamases (4).
Plasmid content and transformation experiments.
Plasmids were extracted from K. pneumoniae FFUL 22K and E. coli transformants by the alkaline lysis method (46). The plasmids isolated from K. pneumoniae FFUL 22K were used to transform E. coli DH5α competent cells with CaCl2, with selection on Luria-Bertani (LB) agar plates containing ceftazidime (10 μg/ml) (GlaxoSmithKline, Lisbon, Portugal).
Cloning experiments.
Plasmid DNA from E. coli DH5α(p22K9) was digested with HindIII (New England Biolabs Inc., Beverly, Mass.), and the fragments were purified from the agarose gel electrophoresis with the Concert rapid gel extraction system (Gibco-BRL Invitrogen Ltd., Paisley, United Kingdom). The purified restriction fragments were ligated to HindIII-linearized pBK-CMV phagemid (Stratagene Europe, Amsterdam, The Netherlands), and the ligation mixture was used to transform E. coli TOP10 One Shot chemically competent cells (Invitrogen Ltd., Paisley, United Kingdom). E. coli TOP10 cells harboring recombinant plasmids were selected on LB agar plates containing kanamycin (30 μg/ml) (Sigma Aldrich Química, Sintra, Portugal) and ampicillin (50 μg/ml) (Sigma Aldrich Química, Sintra, Portugal).
PCR experiments.
Sets of primers were used for detection of blaTEM genes (TEM-A and TEM-B) (4) and amplification of the class 1 integron variable region (5′-CS and 3′-CS) (28) on plasmid p22K9, with Ready-to-go PCR beads (Amersham Biosciences Europe, Lisbon, Portugal). Confirmation of class 3 integrase was done with primers intI3F and intI3R (17) and that of the blaGES-1 gene with primers GES-1A and GES-1B (42). With the published sequences of the intI3 (2, 10) and blaGES/blaIBC genes (15, 16, 29, 40, 41, 42), primers intI3P (5′-AATCCGCTTGCGTTCTGG-3′) and GES-1M (5′-CCAATTTCTCTCAGTAAGAGG-3′) were designed to amplify the promoter region of class 3 integron and the first half of the blaGES-1 gene cassette.
Cloning of PCR amplicons.
For cloning and expression of blaGES-1 gene with the Pc promoter, primers intI3P and GES-1B were used to amplify a 1,388-bp fragment of the class 3 integron. The amplicon was cloned into the SmaI site of pBK-CMV (46), and the ligation mixture was used to transform E. coli TOP10 One Shot chemically competent cells. Transformants harboring recombinant plasmids were selected on LB agar plates containing kanamycin (30 μg/ml) and ceftazidime (50 μg/ml).
DNA sequencing.
DNA sequences were determined on both strands by the dideoxy chain termination method with the Big Dye terminator cycle sequencing kit (Applied Biosystems, Porto, Portugal) and analyzed with an ABI Prism 310 automatic sequencer (Applied Biosystems, Porto, Portugal). The sequences of the cloned DNA in pMFA-1 and of the natural plasmid p22K9 were determined with published or laboratory-designed sequencing primers (gene walking). The nucleotide and the deduced protein sequences were analyzed with the Blast (1) and Clustal W (51) programs, as previously described (12).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper are available in the GenBank/EMBL/DDBJ sequence databases under accession number AY219651.
RESULTS
K. pneumoniae FFUL 22K was isolated in April 1999 at the Hospital Santa Maria (Lisbon, Portugal) from the urine of a patient hospitalized in an intensive care unit with respiratory problems.
K. pneumoniae was resistant to amino- and carboxy-penicillins and associations with clavulanic acid, cefoxitin, cefuroxime, ceftazidime, cefpirome, gentamicin, kanamicin, netilmicin, nalidixic acid, and fluoroquinolones. It showed intermediate susceptibility to cefotaxime, aztreonam, ceftriaxone and amikacin, and was susceptible to cefepime, and imipenem (13). Synergies were observed between clavulanic acid-amoxicillin and cefotaxime, aztreonam, and cefepime, suggesting the production of an extended-spectrum β-lactamase.
Localization and transfer of β-lactamase gene.
Since acquired resistance determinants in K. pneumoniae are often carried on plasmids, the presence of plasmid DNA was investigated by the alkaline lysis method. Agarose gel electrophoresis revealed the presence of three plasmids, ca. 9.4, 6.3, and 3 kb.
Conjugation experiments were not attempted because K. pneumoniae FFUL 22K was resistant to the selective markers of the E. coli strains available at our laboratory. The plasmid preparation was used to transform E. coli DH5α. After selection on LB agar plates with ceftazidime (10 μg/ml), an E. coli transformant that harbored the natural 9.4-kb plasmid, named p22K9, was obtained.
E. coli DH5α(p22K9) displayed a resistance phenotype similar to that of the K. pneumoniae strain except that it was susceptible to ticarcillin-clavulanic acid, cefoxitin, cefotaxime, aztreonam, ceftriaxone, gentamicin, netilmicin, and amikacin (data not shown). Also, potentiation of cefotaxime and aztreonam activity by clavulanate was observed.
Isoelectric focusing showed production of a β-lactamase with an apparent isoelectric point of 5.9 in both K. pneumoniae FFUL 22K and the transformant strain. A second enzyme with a pI of 7.6 was also produced by K. pneumoniae, most probably representing a SHV-1 chromosomal β-lactamase.
Cloning and sequencing of β-lactamase gene.
Preliminary PCR-based experiments failed to detect blaTEM genes and class 1 integrons in the E. coli DH5α transformant.
Plasmid DNA from E. coli DH5α(p22K9) was digested with restriction endonuclease HindIII and ligated to pBK-CMV. The ligation product was transformed into E. coli TOP10, and recombinant clones obtained after selection on kanamycin- and ampicillin-containing plates were analyzed. Restriction analysis of the recombinant plasmid pMFA-1 revealed a 1.2-kb insert, and unlike the parental strain, E. coli TOP10(pMFA-1) showed resistance to aminopenicillins.
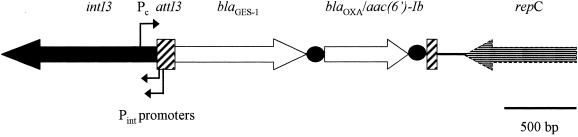
Analysis of the nucleotide sequence from the 1,212-bp insert in pMFA-1 revealed an 864-bp-long open reading frame encoding GES-1 β-lactamase. Also in this case, blaGES-1 appeared to be part of a mobile gene cassette inserted into a class 3 integron-like structure that was named In3-p22K9 (Fig. 1; see below for integron and cassette descriptions).
FIG. 1.
Schematic representation of class 3 integron In3-p22K9 and its flanking region from p22K9. Open reading frames are indicated by arrows, and the 59-base elements are indicated by black solid circles.
Molecular characterization of class 3 integron.
To further analyze the structure of In3-p22K9, gene walking was performed on the natural plasmid p22K9. The class 3 integron was 2,863 bp long and consisted of an intI3 integrase gene, two (Pc and Pint) promoter regions, an attI3 recombination site, a blaGES-1 gene cassette, and a fused blaOXA-10-type/aac(6′)-Ib gene cassette (Fig. 1).
The intI3 gene encodes an integrase that is 98.8% identical to the IntI3 from S. marcescens AK9373 (2, 10). Four substitutions in the deduced amino acid sequences were observed: Gly-6→Arg, Ser-7→Asn, Ala-8→Asp, and Ser-34→Ile. None of them falls in the residues that are highly conserved among the tyrosine family of recombinases neither in the IntI patch (31, 37).
The sequence of the putative Pint promoter nearest the intI3 gene (regions −35[TTGAAA] and −10[CTTACT]) differed in the −10 region by an A/T transversion from the S. marcescens AK9373 integron (−10[CATACT]) (10). The putative Pc sequence (regions −35[TAGACA] and −10[TAGGAT]) differed by a C/A transversion in the −10 region from the reported sequence (−10[TAGGCT]).
The 131-bp region where the 5′ region of attI3 site was located (10) differed by five nucleotides, including two A/T transversions (one of them on Pint) and three transitions (one G/A and two C/T, one of these on intI3 and the other on the simple-site region). None of the substitutions falls in the five GTTRRRY motifs (8, 38).
The gene cassettes were inserted in tandem in the variable region of In3-p22K9. The first one was the blaGES-1 gene cassette, with 1,020 bp and 100% homologous to the gene cassette from P. aeruginosa Pa695 (15). The sequence of the blaGES-1 gene differed by a single silent mutation from that described in K. pneumoniae ORI-1 (42). The amino acid sequence of GES-1 differed by two substitutions from IBC-1, reported in Enterobacter cloacae strains (16, 26), and by one substitution from GES-2 and IBC-2, reported in P. aeruginosa strains (29, 40, 41).
The second gene cassette was 635 bp long and revealed homology with sections of blaOXA-10-type and aac(6′)-Ib gene cassettes. Thus, this cassette was the result of a fusion event between two gene cassettes (Fig. 2). The first 32 bp were 100% homologous to the initial region of blaOXA-10-type gene cassettes (3, 18, 24, 32, 33, 39, 48). The remaining 603 bp, which included the 59-base element, showed 100% homology with previously described aac(6′)-Ib cassettes (27, 32, 39, 42, 43) and differed by a C/A transversion from the aac(6′)-Ib (also known as aacA4) gene cassette present in the integron from S. marcescens AK9373 (10).
FIG. 2.
Sequence alignment and comparison of fusion point of blaOXA-10-type/aac(6′)-Ib fused gene cassette (shown in capital letters) in p22K9 with the initial regions of the blaOXA-10 (24) and aac(6′)-Ib (39) gene cassettes. The nucleotide sequences of the blaOXA-10 and aac(6′)-Ib gene cassettes presenting 100% homology with the fused cassette are also shown in capital letters, where colons indicate identical nucleotides. The putative ribosome-binding sites are underlined, and the translational start codons are double underlined.
The fused blaOXA-10-type/aac(6′)-Ib gene cassette showed a 555-bp open reading frame encoding a 184-amino-acid fusion protein that contained the four conserved motifs of the AAC(6′)-Ib enzymes (50). The deduced amino acid sequence showed three alterations when compared with AAC(6′)-Ib enzymes (27, 39, 43): Thr-2→Lys, Asn-3→Thr, and Ser-4→Phe. These enzymes conferred resistance to gentamicin, while the new enzyme conferred resistance to kanamycin.
The core site (GTTAGCC) of the fused cassette was the same identified in the blaOXA-10-type cassettes (3, 18, 24, 32, 33, 39, 48). The inverse core site (GCCTAAC), which showed a 1-bp mismatch, was 100% homologous to that found in the aac(6′)-Ib cassettes as well the 59-base element (10, 27, 32, 39, 43).
The sequence immediately beyond the second gene cassette was related to the remainder of the original attI3 site. Thereafter, only a region of about 40 bp was clearly related to the end of the integron from S. marcescens AK9373.
Downstream the integron In3-p22K9 and separated by approximately 150 bp, there was a putative open reading frame whose product was 76% identical and 86% similar to the replication protein C of plasmid RSF1010 (47).
Cloning of β-lactamase gene with Pc region and antibiotic susceptibility.
As the insert in pMFA-1 contains only the −10 region of the Pc promoter, since the HindIII restriction endonuclease recognizes and cleaves between the −35 and −10 regions of this promoter, a 1,388-bp fragment of the class 3 integron containing the Pc promoter and the blaGES-1 gene was amplified for cloning and expression. Sequence analysis of the recombinant plasmid pMFA-2 revealed 100% homology with the same sequence determined in the integron.
The MICs of selected β-lactams for K. pneumoniae FFUL 22K and the transformant strains harboring either the natural plasmid p22K9 or the recombinant plasmid pMFA-2 are shown in Table 2. In all cases, the cefuroxime and ceftazidime MICs were higher than those of cefotaxime and aztreonam. All strains were susceptible to imipenem, with similar MICs.
TABLE 2.
β-Lactam susceptibilities of K. pneumoniae FFUL 22K, E. coli DH5α(p 22K9), and E. coli TOP10(pMFA-2)
| β-Lactam | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| K. pneumoniae FFUL 22K | E. coli DH5α(p22K9)a | E. coli DH5αb | E. coli TOP10(pMFA-2)c | E. coli TOP10(pBK-CMV)d | |
| Amoxicillin | >256 | >256 | 4 | >256 | 2 |
| Amoxicillin/clavulanic acid | 12 | 12 | 4 | 16 | 2 |
| Cefuroxime | 256 | >256 | 3 | >256 | 4 |
| Cefotaxime | 6 | 2 | 0.064 | 8 | 0.125 |
| Cefotaxime/clavulanic acid | >1 | 0.19 | NDe | >1 | ND |
| Ceftazidime | 256 | 96 | 0.25 | >256 | 0.75 |
| Ceftazidime/clavulanic acid | >4 | 1 | ND | >4 | ND |
| Aztreonam | 3 | 0.75 | 0.125 | 1.5 | 0.094 |
| Cefepime | 2 | 0.5 | 0.047 | ND | 0.047 |
| Imipenem | 0.25 | 0.19 | 0.19 | 0.38 | 0.25 |
E. coli DH5α (p22K9) expressed β-lactamase GES-1.
The susceptibility of E. coli DH5α is shown for comparison.
E. coli TOP10 (pMFA-2) expressed β-lactamase GES-1.
The susceptibility of E. coli TOP10 carrying the empty vector is also shown for comparison.
ND, not determined.
DISCUSSION
This work was initiated with the observation that K. pneumoniae FFUL 22K exhibited resistance to cefuroxime and ceftazidime and had a positive double-disk synergy test. The apparent pI of 5.9 of the β-lactamase differed from that previously described (pI of 5.6) in K. pneumoniae strains producing the TEM-10 β-lactamase isolated in the same hospital since 1991 (4, 14). Also, the K. pneumoniae strain showed an aztreonam MIC (3.0 μg/ml) lower than those previously determined for the K. pneumoniae strains producing TEM-10, whose aztreonam MICs ranged from 16 to >256 μg/ml (4).
blaGES-1 has been found previously in K. pneumoniae and P. aeruginosa strains isolated at two French hospitals (15, 42) and is located on a gene cassette inserted in class 1 integrons. The integron from K. pneumoniae ORI-1 was located on a 140-kb nontransferable plasmid (42), while that of P. aeruginosa Pa695 was chromosomally located (15). The other blaGES/blaIBC gene cassettes are also inserted in class 1 integrons, located either on large transferable plasmids (blaIBC-1 and blaGES-2) (16, 40, 41) or in the chromosome (blaIBC-2) (29). In the present study, the blaGES-1 gene cassette was inserted at the attI3 site of a class 3 integron, located on a 9.4-kb plasmid. This fact suggests a higher heterogeneity of the genetic environment where blaGES genes can be found.
To date, only a single representative class 3 integron has been reported and characterized (2, 10), although intI3-specific fragments had previously been amplified in blaIMP-positive strains (49) and were detected by DNA-DNA hybridization in several Enterobacteriaceae isolates (17). The integron from S. marcescens AK9373 was located on a ca. 120-kb transferable plasmid, contained blaIMP-1 and aac(6′)-Ib gene cassettes, and was probably part of a transposable element related to Tn402 (2, 10, 25).
The integron In3-p22K9 from K. pneumoniae FFUL 22K was located on a 9.4-kb plasmid and contained a blaGES-1 gene cassette and a fused blaOXA-10-type/aac(6′)-Ib gene cassette. The integron module, consisting of an intI3 allele, an attI3 site and a Pc promoter region, was configured in the same way as the class 3 integron previously characterized (10). Despite the fact that a 21-bp sequence in the terminal region of the integron was 80.9% identical to the sequence of Tn402 and 90.4% to that of S. marcescens AK9373, it differed from both by the absence of IRi, suggesting that probably In3-p22K9 is not part of a transposon backbone.
The IntI3 enzyme from In3-p22K9 differed by four substitutions from the deduced amino acid sequence of the previously characterized IntI3. Since none of these alterations occurred in the conserved residues of the integrases (31, 37) and the IntI3 and IntI1 show stringent specificity for recombination involving the cognate attI site (9), we can assume that this intI3 allele codifies a functional IntI3 integrase.
Among class 1 integrons, four versions of Pc have been identified, which were classified according to their activity (11, 45). In3-p22K9 showed a second version of Pc for class 3 integrons, differing by a C/A transversion in the −10 region from the previously reported version (10). The −35 region of the class 3 promoter (TAGACA) differs from the strong promoter of class 1 integrons (TTGACA) by a T/A transversion at the second nucleotide. Previously, the promoter of a plasmid (pRMH262) containing this mutation showed an intermediate activity, as opposed to the strong activity of the promoter from pRMH280, whose sequence was −35(TTGACA) (11). To ascertain if the C/A transversion influences the expression of the inserted gene cassettes, as previously determined for other mutations in the Pc promoters of class 1 integrons, further studies are needed.
Cassette fusion may occur by a deletion event with endpoints in two adjacent gene cassettes, resulting in the truncation of one or both genes, or by loss of the 59-base element from one cassette, resulting in retention of both gene coding regions (45). In the present study we described the first fused cassette containing truncated versions of blaOXA-10-type and aac(6′)-Ib genes. Although several start codons have been proposed for the aac(6′)-Ib genes (6), the fusion of the blaOXA-10-type and aac(6′)-Ib cassettes placed a translational start codon (ATG) in frame for the aac(6′)-Ib gene, resulting in a fused AAC(6′)-Ib enzyme that conferred resistance to kanamycin. Similarly, in pSTI1 the aac(6′)-Ib gene was fused with the 5′ end of blaOXA-1, resulting in a fused 188-amino-acid protein (6).
This is the first time that a class 3 integron has been found and characterized in Europe, suggesting wide dissemination of integrons belonging to this class. However, it was not found in strains collected during the same period at Hospital Santa Maria, namely Escherichia coli, Enterobacter cloacae, and nonclonal K. pneumoniae strains.
In summary, the molecular characterization of the extended-spectrum β-lactamase gene carried by K. pneumoniae FFUL 22K revealed the presence of a blaGES-1 gene cassette inserted into a class 3 integron, carried by a 9.4-kb plasmid (p22K9). Analysis of the second gene cassette revealed that it was a new fusion cassette that encoded an AAC(6′)-Ib enzyme resulting from translational fusion between a blaOXA-10-type and an aac(6′)-Ib gene cassette.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubert, D., L. Poirel, A. B. Ali, F. W. Goldstein, and P. Nordmann. 2001. OXA-35 is an OXA-10-related β-lactamase from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 48:717-721. [DOI] [PubMed] [Google Scholar]
- 4.Barroso, H., A. Freitas-Vieira, L. M. Lito, J. Melo Cristino, M. J. Salgado, H. Ferreira Neto, J. C. Sousa, G. Soveral, T. Moura, and A. Duarte. 2000. Survey of Klebsiella pneumoniae producing extended-spectrum β-lactamases at a Portuguese hospital: TEM-10 as the endemic enzyme. J. Antimicrob. Chemother. 45:611-616. [DOI] [PubMed] [Google Scholar]
- 5.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casin, I., B. Hanau-Berçot, I. Podglajen, H. Vahagoblu, and E. Collatz. 2003. Salmonella enterica serovar Typhimurium blaPER-1-carrying plasmid pSTI1 encodes an extended-spectrum aminoglycoside 6′-N-acetyltransferase of type Ib. Antimicrob. Agents Chemother. 47:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, C. A., L. Purins, P. Kaewrakon, T. Focareta, and P. A. Manning. 2000. The Vibrio cholerae O1 chromosomal integron. Microbiology 146:2605-2612. [DOI] [PubMed] [Google Scholar]
- 8.Collis, C. M., M.-J. Kim, H. W. Stokes, and R. M. Hall. 1998. Binding of the purified integron DNA integrase IntI1 to integron- and cassette-associated recombination sites. Mol. Microbiol. 29:477-490. [DOI] [PubMed] [Google Scholar]
- 9.Collis, C. M., M.-J. Kim, H. W. Stokes, and R. M. Hall. 2002. Integron-encoded IntI integrases preferentially recognize the adjacent cognate attI site in recombination with a 59-be site. Mol. Microbiol. 46:1415-1427. [DOI] [PubMed] [Google Scholar]
- 10.Collis, C. M., M.-J. Kim, S. R. Partridge, H. W. Stokes, and R. M. Hall. 2002. Characterization of the class 3 integron and the site-specific recombination system it determines. J. Bacteriol. 184:3017-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Da Silva, G. J., M. Correia, C. Vital, G. Ribeiro, J. C. Sousa, R. Leitão, L. Peixe, and A. Duarte. 2002. Molecular characterization of blaIMP-5, a new integron-borne metallo-β-lactamase gene from an Acinetobacter baumanii nosocomial isolate in Portugal. FEMS Microbiol. Lett. 215:33-39. [DOI] [PubMed] [Google Scholar]
- 13.Duarte, A., F. Boavida, F. Grosso, M. Correia, L. M. Lito, J. M. Cristino, and M. J. Salgado. 2003. Outbreak of GES-1 β-lactamase-producing multidrug-resistant Klebsiella pneumoniae in a university hospital in Lisbon, Portugal. Antimicrob. Agents Chemother. 47:1481-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duarte, A., N. Faria, T. Conceição, M. Correia, L. M. Lito, J. M. Cristino, M. J. Salgado, and R. Tenreiro. 2002. Identification of TEM-10 β-lactamase in a Kluyvera sp. and other Enterobacteriaceae at a Portuguese hospital. Antimicrob. Agents Chemother. 46:4041-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubois, V., L. Poirel, C. Marie, C. Arpin, P. Nordmann, and C. Quentin. 2002. Molecular characterization of a novel class 1 integron containing blaGES-1 and a fused product of aac(3)-Ib/aac(6′)-Ib′ gene cassettes in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giakkoupi, P., L. S. Tzouvelekis, A. Tsakris, V. Loukova, D. Sofianou, and E. Tzelepi. 2000. IBC-1, a novel integron-associated class A β-lactamase with extended-spectrum properties produced by an Enterobacter cloacae clinical strain. Antimicrob. Agents Chemother. 44:2247-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein, C., M. D. Lee, S. Sanchez, C. Hudson, B. Phillips, B. Register, M. Grady, C. Liebert, A. O. Summers, D. G. White, and J. J. Maurer. 2001. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45:723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, L. M., D. M. Livermore, G. Dur, M. Akova, and H. E. Akalin. 1993. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37:1637-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 20.Hall, R. M., D. E. Brookes, and H. W. Stokes. 1991. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination crossover point. Mol. Microbiol. 5:1941-1959. [DOI] [PubMed] [Google Scholar]
- 21.Hall, R. M., and H. W. Stokes. 1993. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica 90:115-132. [DOI] [PubMed] [Google Scholar]
- 22.Hansson, K., L. Sundström, A. Pelletier, and P. H. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochhut, B., Y. Lotfi, D. Mazel, S. H. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huovinen, P., S. Huovinen, and G. A. Jacoby. 1988. Sequence of PSE-2 beta-lactamase. Antimicrob. Agents Chemother. 32:134-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, H., Y. Arakawa, S. Ohsuka, R. Wacharoratayankun, N. Kato, and M. Ohta. 1995. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob. Agents Chemother. 39:824-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kartali, G., E. Tzelepi, S. Pournaras, C. Kontopoulou, F. Kontos, D. Sofianou, A. N. Maniatis, and A. Tsakris. 2002. Outbreak of infections caused by Enterobacter cloacae producing the integron-associated β-lactamase IBC-1 in a neonatal intensive care unit of a Greek hospital. Antimicrob. Agents Chemother. 46:1577-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, M. D., S. Sanchez, M. Zimmer, U. Idris, M. E. Berrang, and P. F. McDermott. 2002. Class 1 integron-associated tobramycin-gentamicin resistance in Campylobacter jejuni isolated from the broiler chicken house environment. Antimicrob. Agents Chemother. 46:3660-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mavroidi, A., E. Tzelepi, A. Tsakris, V. Miriagou, D. Sofianou, and L. S. Tzouvelekis. 2001. An integron-associated β-lactamase (IBC-2) from Pseudomonas aeruginosa is a variant of the extended-spectrum β-lactamase IBC-1. J. Antimicrob. Chemother. 48:627-630. [DOI] [PubMed] [Google Scholar]
- 30.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 1998. A Distinctive class of integron in the Vibrio cholera genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 31.Messier, N., and P. H. Roy. 2001. Integron integrases possess a unique additional domain necessary for activity. J. Bacteriol. 183:6699-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mugnier, P., I. Casin, A. T. Bouthors, and E. Collatz. 1998. Novel OXA-10-derived extended-spectrum β-lactamases selected in vivo or in vitro. Antimicrob. Agents Chemother. 42:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mugnier, P., I. Podglajen, F. W. Goldstein, and E. Collatz. 1998. Carbapenems as inhibitors of OXA-13, a novel, integron-encoded β-lactamase in Pseudomonas aeruginosa. Microbiology 144:1021-1031. [DOI] [PubMed] [Google Scholar]
- 34.Naas, T., L. Poirel, A. Karim, and P. Nordmann. 1999. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 176:411-419. [DOI] [PubMed] [Google Scholar]
- 35.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Committee for Clinical Laboratory Standards. 2000. Methods for disk antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 37.Nield, B. S., A. J. Holmes, M. R. Gillings, G. D. Recchia, B. C. Mabbutt, K. M. H. Nevalainen, and H. W. Stokes. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195:59-65. [DOI] [PubMed] [Google Scholar]
- 38.Partridge, S. R., G. D. Recchia, C. Scaramuzzi, C. M. Collis, H. W. Stokes, and R. M. Hall. 2000. Definition of the attI1 site of class 1 integrons. Microbiology 146:2855-2864. [DOI] [PubMed] [Google Scholar]
- 39.Poirel, L., D. Girlich, T. Naas, and P. Nordmann. 2001. OXA-28, an extended-spectrum variant of OXA-10 β-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob. Agents Chemother. 45:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poirel, L., G. F. Weldhagen, C. de Champs, and P. Nordmann. 2002. A nosocomial outbreak of Pseudomonas aeruginosa isolates expressing the extended-spectrum β-lactamase GES-2 in South Africa. J. Antimicrob. Chemother. 49:561-565. [DOI] [PubMed] [Google Scholar]
- 41.Poirel, L., G. F. Weldhagen, T. Naas, C. de Champs, M. G. Dove, and P. Nordmann. 2001. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45:2598-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poirel, L., I. Le Thomas, T. Naas, A. Karim, and P. Nordmann. 2000. Biochemical-sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poirel, L., T. Lambert, S. Türkoglü, E. Ronco, J.-L. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Scholz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271-288. [DOI] [PubMed] [Google Scholar]
- 48.Scoulica, E., A. Aransay, and Y. Tselentis. 1995. Molecular characterization of the OXA-7 β-lactamase gene. Antimicrob. Agents Chemother. 39:1379-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shmara, A., N. Weinsetel, K. J. Dery, R. Chavideh, and M. E. Tolmasky. 2001. Systematic analysis of a conserved region of the aminoglycoside 6′-N-acetyltransferase type Ib. Antimicrob. Agents Chemother. 45:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]