Abstract
The multiresistance transposon Tn1331, which mediates resistance to several aminoglycosides and β-lactams, includes the aac(6′)-Ib, aadA1, blaOXA-9, and blaTEM-1 genes. The nucleotide sequence of aac(6′)-Ib includes a region identical to that of the blaTEM-1 gene. This region encompasses the promoter and the initiation codon followed by 15 nucleotides. Since there were three possible translation initiation sites, the amino acid sequence at the N terminus of the aminoglycoside 6′-N-acetyltransferase type Ib [AAC(6′)-Ib] was determined and was found to be SIQHF. This result indicated that aac(6′)-Ib includes a translational fusion: the first five amino acids of the leader peptide of the TEM β-lactamase are fused to the rest of the AAC(6′)-Ib protein. This gene fusion could have formed during the genesis of Tn1331 as a consequence of the generation of a 520-nucleotide duplication (M. E. Tolmasky, Plasmid 24:218-226, 1990). An identical gene isolated from a Serratia marcescens strain has been previously described (G. Tran van Nhieu and E. Collatz, J. Bacteriol. 169:5708-5714, 1987). Extraction of the periplasmic proteins of E. coli harboring aac(6′)-Ib by spheroplast formation showed that most of the AAC(6′)-Ib protein is present in the cytoplasm. A genetic fusion to phoA confirmed these results. AAC(6′)-Ib was shown to be evenly distributed inside the cell's cytoplasm by fluorescent microscopy with an AAC(6′)-Ib-cyan fluorescent protein fusion.
Bacterial resistance to aminoglycosides in the clinical setting is often due to three major groups of modifying enzymes: acetyltransferases, nucleotidyltransferases, and phosphotransferases (18, 31, 50). The modifications of the antibiotic molecule mediated by these enzymes prevent the aminoglycosides from exerting their biological activity. Recent studies led to considerable advances in the genetics, structure, biochemistry, and mechanisms of dissemination of several aminoglycoside-modifying enzymes (8, 13, 15, 24, 32, 38, 40-42, 45, 48, 51). On the other hand, there have been only limited efforts to determine their subcellular location (reviewed by Shaw et al. [31]). While some of them seem to be located in the cytoplasm, some others seem to be periplasmic (25, 46). However, many of these studies were performed using methods of physical fractionation of cellular compartments such as osmotic shock, which has been recently shown to generate a molecular sieve by transient damage of the bacterial envelope, permitting the release of small proteins regardless of their subcellular location (44). Furthermore, information about the distribution of the protein in a given cell compartment is lacking.
The aminoglycoside 6′-N-acetyltransferase type Ib [AAC(6′)-Ib] belongs to the GCN5-related N-acetyltransferase superfamily (21) and confers resistance to several aminoglycosides including kanamycin (KAN), amikacin (AMK), and tobramycin. This protein has been recently characterized by random and alanine scanning mutagenesis (8, 24, 28, 32) as well as by analysis of natural variants (6, 7). As part of the characterization of this enzyme we determined its N-terminal amino acid sequence as well as its subcellular localization and distribution by physical fractionation, gene fusion, and fluorescence microscopy.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strains and plasmids are described in Table 1. Plasmid pMET33.6 was generated by inserting the 3-kb pJHCMW1 BamHI fragment, which includes aac(6′)-Ib, into the BamHI site of the expression vector pT7-5 (36) in the proper orientation for transcription of aac(6′)-Ib from the T7 promoter. To generate a fusion between AAC(6′)-Ib and a version of alkaline phosphatase that lacks the signal peptide, an amplicon containing the aac(6′)-Ib gene flanked by XbaI and BamHI restriction sites was generated using the following primers: 5′-GCTCTAGAAGGGCCTCGTGATACGCC (A corresponds to nucleotide 7107 in the sequence deposited under accession number AF479774) and 5′-CGCGGATCCTTCGCTCGAATGCCTGGCG (G corresponds to nucleotide 7872 in the sequence deposited under accession number AF479774). The amplicon was inserted in XbaI- and BamHI-digested pBCSK (Stratagene). The resulting recombinant plasmid was then treated with BamHI and XhoI and ligated to the 2.4-kb BamHI-XhoI fragment containing the alkaline phosphatase gene obtained from pUCH2 to generate pKDR2PEP. Plasmids pMS2B6 and pMS27ss were used as controls in alkaline phosphatase activity experiments. E. coli(pMSB26) produces significant amounts of alkaline phosphatase. E. coli(pMS27ss) produces hybrid protein but no alkaline phosphatase activity (33). Plasmid pAACCFP was constructed by amplification of the aac(6′)-Ib gene with pJHCMW1 as a template and the primers 5′-GGCCCATGGTGAGTATTCAACATTTCCAAAC (C corresponds to nucleotide 7324 in the sequence deposited under accession number AF479774) and 5′-GCGCTCGAGTGGTACCGGTGGCCCGTGGATCCGAAGTCTGGACATGGCAACACTGCGTGTTCGCTCG (G corresponds to nucleotide 7904 in the sequence deposited under accession number AF479774). To generate an NcoI site, the forward primer was designed containing the GTG sequence between the ATGAGT sequences found in the natural gene. This results in the addition of valine at the N-terminal end of the protein. The enzyme keeps its activity in spite of this modification. The amplicon was digested with NcoI and XhoI and ligated to NcoI- and XhoI-digested pLAU15 (I. Lau and D. Sherratt, unpublished data), a vector constructed by cloning the cyan fluorescent protein (CFP) gene from pSG1190 (14) into pBAD24 (16). The plasmid pAACCFP contains a fusion of the AAC(6′)-Ib protein to the CFP. Plasmids pTGS and pJDT, which contain the TorA leader peptide fused to Ssr-tagged green fluorescent protein (GFP) (12) and a TorA-GFP fusion (37), respectively, were used as controls in microscopy experiments.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic or genotype | Reference or origin |
|---|---|---|
| E. coli strains | ||
| AB1157 | thr-1 araC14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 qsr′ glnV44 galK2 λ−hisG4 rfbD1 mgl-51 rpoS396 rpsL31(Smr) kdgK51 xylA5 mtl-1 argE3 thi-1 supE44 | 2 |
| HB101 | supE44 ara-14 galK2 Δ(gpt-proA)62 lacY1 rpsL31(Stmr) xylA5 mtl-1 recA13 Δ(mcrC-mrr) HsdS− | 5 |
| BL21(DE3)(pLysE) | F−dcm ompT hsdS(rB− mB−) gal λ(DE3) (pLysE Chlr) | 34 |
| Plasmids | ||
| pJHCMW1 | Natural isolate. Carries aac(6′)-Ib | 30 |
| pT7-5 | Expression vector; Ampr | 36 |
| pMET33.6 | 3-kb pJHCMW1 BamHI fragment including aac(6′)-Ib inserted into the BamHI site of pT7-5 | This work |
| pAACCFP | Recombinant plasmid coding for AAC(6′)-Ib-CFP fusion (under the control of the araBAD promoter) in pLAU15 | This work |
| pLAU15 | CFP fusion vector | I. Lau |
| pTGS | TorA leader peptide fused to Ssr-tagged GFP in pBAD33 | 12 |
| pJDT1 | TorA leader peptide fused to GFP in pBAD24 | 37 |
| pKDR2PEP | Recombinant plasmid coding for AAC(6′)-Ib-alkaline phosphatase fusion | This work |
| pUCH2 | A 5.6-kb phoA fusion cloning vector with polylinker sequence replacing signal sequence: Ampr | 33 |
| pMS2B6 | Contains the first 97 amino acids of prepilin (pilA) fused in frame to phoA lacking signal sequence: Ampr | 33 |
| pMS27ss | Contains the first 10 amino acids of prepilin (pilA) fused in frame to phoA lacking signal sequence; Ampr | 33 |
General DNA procedures.
Plasmid DNA preparations were performed using the Qiagen plasmid minikit (Qiagen Inc.). Restriction endonuclease and ligase treatments were carried out as recommended by the supplier (New England Biolabs). Transformation was carried out as described before (9). PCRs were performed using the HotStarTaq DNA polymerase kit (Qiagen).
Determination of the N-terminal sequence of AAC(6′)-Ib.
E. coli BL21(DE3)(pLysE) was transformed with pMET33.6, and the cells were cultured in L broth containing 100 μg of ampicillin/ml at 37°C until the optical density at 600 nm (OD600) was 0.7. At this point IPTG (isopropyl-β-d-galactopyranoside) was added to a concentration of 1 mM to induce protein expression. After incubation for an additional 3 h the cells were harvested by centrifugation. Two-dimensional electrophoresis of a total protein sample was then performed according to the method of O'Farrell (23) at Kendrick Labs, Inc. Isoelectric focusing was carried out in a glass tube (2.0-mm inner diameter) in a pH 3.5 to 10 gradient (Pharmacia). One microgram of tropomyosin (internal standard), which migrates as a doublet with a 33-kDa, pI 5.2 spot, was added to the sample. After isoelectric focusing the tube gel was equilibrated for 10 min in a buffer containing 10% glycerol, 50 mM dithiothreitol, 2.3% sodium dodecyl sulfate (SDS), and 0.0625 M Tris, pH 6.8. Then the tube gel was sealed to the top of a stacking gel that overlaid a 10% acrylamide slab gel (0.75 mm thick). After electrophoresis the gel was placed in transfer buffer (12.5 mM Tris [pH 8.8], 86 mM glycine, 10% methanol) and transferred to a polyvinylidene difluoride membrane overnight at 200 mA. The membrane was stained with Coomassie brilliant blue R-250, and the spot corresponding to AAC(6′)-Ib was cut from the polyvinylidene difluoride membrane and sequenced on an Applied Biosystems (Foster City, Calif.) model 494 protein sequencer at the Protein Chemistry Core Facility, Columbia University.
Cell fractionation.
Extraction of periplasmic proteins was carried out by spheroplast formation (47). E. coli HB101(pJHCMW1) cells were harvested by centrifugation from cultures grown overnight at 37°C, washed, and resuspended in 0.1 volume of spheroplast buffer (100 mM Tris HCl [pH 8.0], 0.5 M sucrose, 0.5 mM EDTA). Then, lysozyme was added to the suspension to a 1-mg/ml concentration followed by addition of 0.2 volume of H2O. After incubation at room temperature for 30 min, 1 M MgCl2 was added to a concentration of 20 mM to stabilize the spheroplasts. This suspension was then centrifuged at 5,000 × g for 5 min at 4°C, and the supernatant containing the periplasmic content was saved. The spheroplasts (pellet) were suspended in a solution containing 1 mM Tris HCl (pH 8.0), 20 mM MgCl2, and 1 mM EDTA; sonicated; and ultracentrifuged at 100,000 × g for 30 min at 4°C. The cytoplasmic proteins were recovered in the supernatant.
Assay of alkaline phosphatase activity.
Alkaline phosphatase was assayed basically as described previously (27) on E. coli HB101 cells harboring either pJHCMW1, pKDR2PEP, pMS27ss (control coding for a cytoplasmic protein), or pMS2B6 (control coding for a periplasmic protein) cultured in L broth (expression of fusion proteins encoded by the plasmids is not regulated by the phosphate concentration). L broth containing either 100 μg of ampicillin/ml (pMS27ss and pMS2B6) or 25 μg of AMK/ml (pJHCMW1 and pKDR2PEP) was inoculated with overnight cultures and incubated at 37°C until it reached an OD600 of 0.6. Cells from 1 ml of each culture were harvested by centrifugation at room temperature and washed twice with ice-cold buffer containing 1 mM iodoacetamide and 1 M Tris-HCl, pH 8. The cells were then resuspended in 3 ml of 0.1 M Tris-HCl, pH 8. Then, 0.1 ml of bacterial suspension was mixed with 0.9 ml of buffer containing 1 M Tris-HCl (pH 8), 1 mM ZnCl2, and 0.001% SDS, followed by addition of 20 μl of chloroform. The suspensions were incubated successively for 5 min at 37°C, 5 min at 0°C, and 1 min at 37°C followed by addition of 100 μl of 4-mg/ml I-nitrophenylphosphate in 1 M Tris-HCl (pH 8) and incubation at 37°C for 10 min. The reaction was stopped by addition of 120 μl of a solution containing 0.5 M EDTA (pH 8) and 1 M KH2PO4 (pH 8) followed by incubation on ice for 5 min. After removal of the cell debris by centrifugation the OD420 and OD550 values were determined. Units of alkaline phosphatase activity were calculated as (OD420 − 1.75 × OD550)/(OD600 × time of the reaction) × 1,000.
Determination of MICs.
MICs were determined by the E-test method (49) with commercial strips (AB Biodisks) according to the procedures recommended by the supplier with the additions to the medium indicated in the text.
Immunoblot analysis.
Immunoblot analysis was carried out basically as described previously (24). Proteins were subjected to SDS-polyacrylamide gel electrophoresis on 12.5% acrylamide gels with prestained molecular weight standards. The proteins were electrophoretically transferred to nitrocellulose paper (0.45-μm pore size; BA85; Schleicher and Schuell, Inc.) with the Trans-Blot cell (Bio-Rad) under the conditions recommended by the supplier. The blots were then blocked for 1 h at room temperature in 5% skim milk-0.05% Triton X-100-0.01% thimerosal, rinsed in NIBB (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 0.05% Triton X-100), and incubated for 1 h at room temperature with either anti-AAC(6′)-Ib serum, monoclonal anti-β-galactosidase (Promega), or anti-β-lactamase serum (4). The blots were the rinsed three times in NIBB and incubated for 1 h in goat anti-rabbit immunoglobulin G (heavy plus light)-alkaline phosphatase conjugate (Bio-Rad). The blots were rinsed three times in a buffer containing 20 mM Tris HCl (pH 7.6), 140 mM NaCl, and 0.1% (vol/vol) Tween 20 and visualized using Supersignal West Pico chemiluminescent substrate (Pierce) according to the directions of the manufacturer.
Microscopy.
For microscopic analysis E. coli AB1157 (35) was transformed with pAACCFP, pTGS (12), or pJDT1 (37) and cultured for 2 h at 37°C before addition of l-arabinose at the indicated concentrations. Expression was allowed to occur for 1 h at 37°C followed by the addition of 300 μg of rifampin/ml and incubation at 30°C for 45 min. In the case of E. coli AB1157(pJDT1), at this moment the cells were harvested, resuspended in 1 ml of L broth, and incubated at 30°C without shaking for 2 h. The published procedure was altered in this fashion to reduce fluorescence because the fusion in this plasmid was done using the redshifted GFPmut3* variant, a more fluorescent form than the wild-type GFP (37). Next, to label the membranes, 200 μg of FM5-95/ml was added to all samples followed by incubation at 37°C with shaking for 15 min. The cells were then immediately examined using filter sets 31044v2 to detect CFP [fused to AAC(6′)-Ib protein as described above], 31019 to detect GFP (fused to control proteins as described above), and 31058 to detect FM5-95 (Chroma Technology Corp., Quebec City, Quebec, Canada). Images were acquired using a cooled charge-coupled device camera (Princeton Instruments) and Metamorph 3.5 image acquisition software (Universal Imaging Corporation) from an Olympus BX50 fluorescence microscope. Final images were transferred to Adobe Photoshop 5.0.
RESULTS AND DISCUSSION
Amino acid sequence of the AAC(6′)-Ib N terminus.
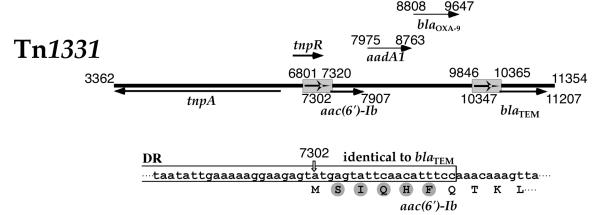
Inspection of the N-terminal region of the Tn1331-carried aac(6′)-Ib gene nucleotide sequence shows that there are at least three potential translation initiation sites at coordinate 7302, 7389, or 7410 according to the numbering in the sequence deposited in GenBank (accession number AF479774) (30). Besides, the protein could undergo further processing before becoming the mature form of the enzyme. Therefore, we determined the amino acid sequence of the AAC(6′)-Ib N terminus. Edman degradation of the protein indicated that the first five amino acids are SIQHF, a result consistent with initiation of translation at nucleotide 7302 (30) and no further processing. Inspection of the nucleotide sequence of the aac(6′)-Ib included in Tn1331 indicates that there is a natural gene fusion of a sequence identical to the promoter and the first six codons of blaTEM with the rest of the structural aac(6′)-Ib gene (22, 39). An identical fusion isolated from a Serratia marcescens strain has been previously described (43). This fusion could have formed during the genesis of Tn1331 as a consequence of the generation of a 520-bp duplication which encompasses a portion of tnpR and the beginning of the blaTEM gene (Fig. 1) (30, 39). While the five amino acids SIQHF belong to the 23-amino-acid leader peptide in the TEM β-lactamase (17, 26) and are therefore removed after translocation to the periplasmic space, they are part of the mature AAC(6′)-Ib amino acid sequence. Casin et al. recently proposed that, due to the diversity of rearrangements that are associated with the insertion of aac(6′)-Ib gene cassettes, insertion may result in generation of variants of the gene at the N-terminal end (6). These authors suggested that the predominance of this gene among the family Enterobacteriaceae could be due to flexible structural requirements for the AAC(6′)-Ib N terminus. We are currently generating mutants with modified N-terminal ends to better understand the requirements at the N terminus for proper folding and activity of AAC(6′)-Ib.
FIG. 1.
Genetic map of Tn1331. The genetic map on top shows the relevant genes with the coordinates as defined in the sequence deposited in GenBank (accession number AF479774). The gray boxes encompassing nucleotides 6801 to 7320 and 9846 to 10365 represent the 520-bp direct repeats. The arrows indicate the locations of some genes [tnpA, tnpR, aac(6′)-Ib, and blaTEM-1]. The translation initiation codon of aac(6′)-Ib is at coordinate 7302, and that of blaTEM-1 is at coordinate 10347. The nucleotide sequence shown below the genetic map depicts the end of the direct repeat (open box), which is identical to the corresponding region in the blaTEM-1 gene. The amino acid sequence is shown below the nucleotide sequence. Gray circles show the amino acids identified by sequencing of the N terminus of AAC(6′)-Ib.
Physical separation of periplasmic and cytoplasmic proteins.
Protein translocation across the cytoplasmic membrane of gram-negative bacteria can be achieved by a number of mechanisms such as the general Sec pathway (11), the alternative targeting route to the cytoplasmic membrane mediated by the signal recognition particle-dependent pathway (20), the YidC-dependent process (29), and the twin arginine translocation system (10). We did not find a leader peptide or the signature characteristic of Tat-translocated proteins (12) in AAC(6′)-Ib. However, since the presence of these signals may not be absolutely required for translocation (20) and information about the distribution of aminoglycoside-modifying enzymes in the cell is not available, we determined the location and distribution of AAC(6′)-Ib in E. coli cells.
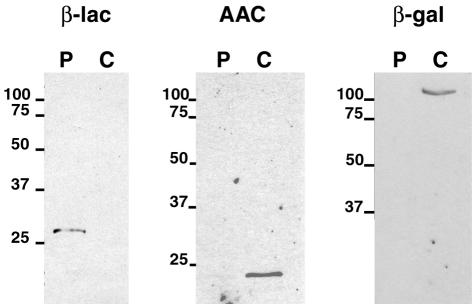
Extraction of the periplasmic proteins of E. coli(pJHCMW1) by spheroplast formation, a technique that includes an osmotic shock step, followed by immunoblot analysis with anti-AAC(6′)-Ib serum, showed that most of the AAC(6′)-Ib signal is present in the extract obtained from inside the cells (Fig. 2). As controls identical immunoblotting assays were carried out using either anti-β-lactamase (a periplasmic protein) serum or monoclonal anti-β-galactosidase (a cytoplasmic protein). As expected, the controls were detected in the periplasmic (β-lactamase) or cytoplasmic (β-galactosidase) protein extract (Fig. 2). These results indicated that AAC(6′)-Ib is located within the cytoplasm of the cells. However, when cell fractionation assays were carried out using harsher osmotic shock conditions, no β-galactosidase was detected in the periplasmic space but a considerable fraction of the total AAC(6′)-Ib was found in this compartment (data not shown). This could be due to the recent finding that, in osmotically shocked E. coli, proteins are released through a molecular sieve formed by the damaged cell envelope, making the osmotic shock separating technique a less than perfect method to extract periplasmic proteins (44). Cytoplasmic proteins small in native size tend to be released after osmotic shock treatment while larger proteins or protein complexes remain inside the cells (44). Therefore, results obtained by physical fractionation of cells should be confirmed by other methods.
FIG. 2.
Immunological detection of AAC(6′)-Ib, β-lactamase, and β-galactosidase in periplasmic and cytoplasmic fractions. The cytoplasmic and periplasmic protein content of E. coli(pJHCMW1) cells was obtained as described in Materials and Methods. Equal amounts of proteins were separated by SDS-polyacrylamide gel electrophoresis and subjected to immunoblotting with anti-AAC(6′)-Ib serum (AAC), anti-β-lactamase serum (β-lac), or monoclonal anti-β-galactosidase (β-gal). Numbers at left of panels are molecular masses in kilodaltons.
Genetic fusion AAC(6′)-Ib-PhoA.
To confirm the location of the Tn1331-encoded AAC(6′)-Ib we used a genetic fusion to alkaline phosphatase, which is active only when it is located in the periplasmic space. Taking advantage of this property, several gene fusions to phoA have been generated to characterize a variety of protein properties such as their transmembrane nature, identification of periplasmic regions of inner membrane proteins, or export signals (1, 19, 33). A gene fusion encoding AAC(6′)-Ib-PhoA was constructed as described in Materials and Methods (plasmid pKDR2PEP), and after determination that the fusion protein was functional (E. coli cells carrying this recombinant plasmid were resistant to AMK and KAN) the alkaline phosphatase activity in cells carrying pKDR2PEP was determined. As controls E. coli cells were also transformed with either pMS2B6 or pMS27ss, which encode fusion proteins that are located in the periplasm and cytoplasm, respectively. With the alkaline phosphatase activity of extracts from E. coli(pMS2B6) cells being set at 1, the relative activities of cells carrying pMS27ss, pKDR2PEP, and pJHCMW1 were 0.2, 0.1, and 0.1, respectively. Thus, no alkaline phosphatase activity was detected in cells carrying pKDR2PEP. This was also the case for E. coli cells harboring pMS27ss or pJHCMW1 (which does not encode a protein fusion). On the other hand cells carrying pMS2B6 showed strong activity. The results of this experiment are in agreement with those obtained by physical separation of the periplasmic and cytoplasmic content. These assays were carried out using E. coli HB101 as the host. Since this strain contains a chromosomal alkaline phosphatase gene, a control experiment was carried out to confirm that no activity was produced in the culture conditions used. Downregulation by the levels of phosphate present in the culture medium has been proposed as the cause of this lack of activity (33). Conversely, expression of the fusion proteins encoded by the plasmid constructs is not regulated by the phosphate concentration of the medium (33).
Genetic fusion AAC(6′)-Ib-CFP.
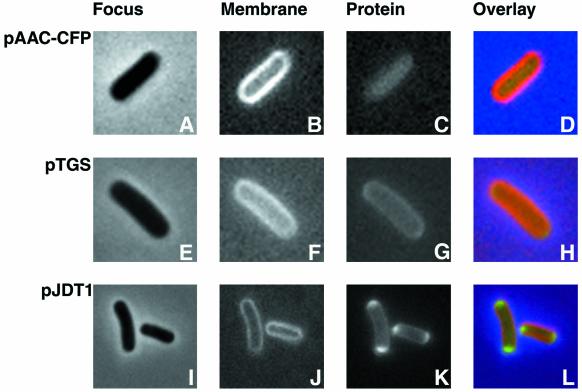
Although the experiments described above gave a strong indication that AAC(6′)-Ib is located in the cytoplasm of E. coli cells, they did not provide information about the distribution of the protein in this compartment. Therefore, we carried out an experiment that permits the visualization of the protein. To directly visualize the localization of the aminoglycoside AAC(6′)-Ib in E. coli cells, we generated the plasmid pAACCFP, which includes a fusion between aac(6′)-Ib and the gene encoding the CFP under the control of the araBAD promoter. The plasmid pAACCFP was introduced into E. coli by transformation, and the transformant cells were first tested for resistance to AMK and KAN. Table 2 shows that E. coli AB1157(pAACCFP) becomes resistant to both aminoglycosides in the presence of l-arabinose, indicating that the fusion protein is functional. The MICs increased as the concentration of arabinose was increased, demonstrating that the araBAD promoter was responsible for expression of the fusion protein. To visualize this fusion protein, cells were grown and induced in the presence of l-arabinose followed by observation under the microscope with the appropriate filters to detect membrane structures labeled by FM5-95 or the fluorescent proteins. Figure 3A to D shows that the fluorescence due to the presence of the fused AAC(6′)-Ib protein is confined to the cell's cytoplasm. The colored overlay shows the membrane in red on the outside and the protein in green inside the cytoplasm. Since we have observed previously that proteins with polar localization could be seen evenly distributed in the cytoplasm when they were overexpressed (3), we analyzed the location of the fused protein at different l-arabinose concentrations. The results indicated that, as soon as the protein was visible, its distribution was homogenous in the cytoplasm. Figure 3 also shows the location of control proteins, which are coded for by two plasmids, pTGS and pJDT1. These control fusion proteins are located within the periplasmic space. Plasmid pTGS encodes a periplasmic protein fusion consisting of the TorA (trimethylamine N-oxide reductase) leader peptide fused to SsrA-tagged GFP (12) (Fig. 3E to H). Plasmid pJDT1 carries a gene fusion coding for a protein consisting of the RR-signal peptide of TorA plus four amino acids of the mature protein fused in frame to GFPmut3*, a redshifted variant of GFP that is more fluorescent than the wild type (37). Thomas et al. determined that the protein encoded by pJDT1 was located in periplasmic space accumulating at the ends of the cell (37). Figures 3I to L show this location and distribution in our experiment.
TABLE 2.
Susceptibilities to AMK and KAN of E. coli AB1157(pAACCFP)
| % l-Arabinose | MIC (μg/ml)a
|
|
|---|---|---|
| AMK | KAN | |
| 0 | 0.5 | 1.5 |
| 0.01 | 1.5 | 6 |
| 0.1 | 8 | 64 |
MICs were determined using the E-test method as described in Materials and Methods. Mueller-Hinton agar plates were supplemented with the indicated concentrations of l-arabinose.
FIG. 3.
Visualization of protein fusions by fluorescence microscopy. Cells containing pAACCFP, pTGS, or pJDT1 were treated as described in Materials and Methods. The membranes of all three strains were stained by incubation in 200 μg of FM5-95/ml at 37°C with shaking for 15 min. Cells were focused (A, E, and I) and examined using filter sets 31044v2 to detect CFP (C), 31019 to detect GFP (G and K), and 31058 to detect FM5-95 (B, F, and J). Overlays were generated by coloring the membranes red and the fusion proteins green (D, H, and L).
Conclusions.
The Tn1331-encoded aac(6′)-Ib has a fusion to the initial region of the blaTEM gene at its N-terminal end. Here we show that translation starts at nucleotide 7302 and the protein does not undergo further modification. Therefore, the first five amino acids are identical to those of the leader peptide of the TEM β-lactamase. However, while they are removed with the leader peptide after translocation of β-lactamase, they are part of the mature AAC(6′)-Ib. Physical extraction of proteins from the periplasmic space as well as genetic fusions followed by biochemical and microscopy analysis indicated that the AAC(6′)-Ib protein is located in the cytoplasm and is evenly distributed in this compartment.
Acknowledgments
This work was supported by Public Health grant AI47115-01 (M.E.T.) from the National Institutes of Health and a grant from Wellcome Trust (D.J.S.). D.B., K.J.D., and M.S.L.W. were supported by MSD grant R25 GM56820-03 and LS Basin MIRT T37 TW00048-05 from the National Institutes of Health.
We thank G. Georgiou for generously providing the monoclonal anti-β-lactamase serum and the pTGS plasmid and I. Lau for providing plasmid pLAU15. We are also indebted to C. Robinson for the generous gift of plasmid pJDT1 and S. Lory for the generous gift of plasmids pMS2B6 and pMS27ss.
K.J.D., B.S., and M.S.L.W. contributed equally to the work.
REFERENCES
- 1.Adler, J., and E. Bibi. 2002. Membrane topology of the multidrug transporter MdfA: complementary gene fusion studies reveal a nonessential C-terminal domain. J. Bacteriol. 184:3313-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann, B. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2495. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 3.Barre, F. X., M. Aroyo, S. D. Colloms, A. Helfrich, F. Cornet, and D. J. Sherratt. 2000. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 14:2976-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowden, G. A., and G. Georgiou. 1990. Folding and aggregation of beta-lactamase in the periplasmic space of Escherichia coli. J. Biol. Chem. 265:16760-16766. [PubMed] [Google Scholar]
- 5.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 6.Casin, I., F. Bordon, P. Bertin, A. Coutrot, I. Podglajen, R. Brasseur, and E. Collatz. 1998. Aminoglycoside 6′-N-acetyltransferase variants of the Ib type with altered substrate profile in clinical isolates of Enterobacter cloacae and Citrobacter freundii. Antimicrob. Agents Chemother. 42:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casin, I., B. Hanau-Bercot, I. Podglajen, H. Vahaboglu, and E. Collatz. 2003. Salmonella enterica serovar Typhimurium blaPER-1-carrying plasmid pSTI1 encodes an extended-spectrum aminoglycoside 6′-N-acetyltransferase of type Ib. Antimicrob. Agents Chemother. 47:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chavideh, R., S. Sholly, D. Panaite, and M. E. Tolmasky. 1999. Effects of F171 mutations in the 6′-N-acetyltransferase type Ib [AAC(6′)-Ib] enzyme on susceptibility to aminoglycosides. Antimicrob. Agents Chemother. 43:2811-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, S., A. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalbey, R. E., and C. Robinson. 1999. Protein translocation into and across the bacterial plasma membrane and the plant thylakoid membrane. Trends Biochem. Sci. 24:17-22. [DOI] [PubMed] [Google Scholar]
- 11.Danese, P. N., and T. J. Silhavy. 1998. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet. 32:59-94. [DOI] [PubMed] [Google Scholar]
- 12.DeLisa, M. P., P. Samuelson, T. Palmer, and G. Georgiou. 2002. Genetic analysis of the twin-arginine translocator secretion pathway in bacteria. J. Biol. Chem. 277:29825-29831. [DOI] [PubMed] [Google Scholar]
- 13.Ekman, D. R., E. L. DiGiammarino, E. Wright, E. D. Witter, and E. H. Serpersu. 2001. Cloning, overexpression, and purification of aminoglycoside antibiotic nucleotidyltransferase (2")-Ia: conformational studies with bound substrates. Biochemistry 40:7017-7024. [DOI] [PubMed] [Google Scholar]
- 14.Feucht, A., and P. J. Lewis. 2001. Improved plasmid vectors for the production of multiple fluorescent protein fusions in Bacillus subtilis. Gene 264:289-297. [DOI] [PubMed] [Google Scholar]
- 15.Fong, D. H., and A. M. Berghuis. 2002. Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 21:2323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadonaga, J. T., A. Pluckthun, and J. R. Knowles. 1985. Signal sequence mutants of beta-lactamase. J. Biol. Chem. 260:16192-16199. [PubMed] [Google Scholar]
- 18.Kotra, L. P., J. Haddad, and S. Mobashery. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44:3249-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manting, E. H., and A. J. Driessen. 2000. Escherichia coli translocase: the unravelling of a molecular machine. Mol. Microbiol. 37:226-238. [DOI] [PubMed] [Google Scholar]
- 21.Neuwald, A. F., and D. Landsman. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154-155. [DOI] [PubMed] [Google Scholar]
- 22.Nobuta, K., M. E. Tolmasky, L. M. Crosa, and J. H. Crosa. 1988. Sequencing and expression of the 6′-N-acetyltransferase gene of transposon Tn1331 from Klebsiella pneumoniae. J. Bacteriol. 170:3769-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 24.Panaite, D. M., and M. E. Tolmasky. 1998. Characterization of mutants of the 6′-N-acetyltransferase encoded by the multiresistance transposon Tn1331: effect of Phen171-to-Leu171 and Tyr80-to-Cys80 substitutions. Plasmid 39:123-133. [DOI] [PubMed] [Google Scholar]
- 25.Perlin, M. H., and S. A. Lerner. 1981. Localization of an amikacin 3′-phosphotransferase in Escherichia coli. J. Bacteriol. 147:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pluckthun, A., and J. R. Knowles. 1987. The consequences of stepwise deletions from the signal-processing site of beta-lactamase. J. Biol. Chem. 262:3951-3957. [PubMed] [Google Scholar]
- 27.Pourcher, T., E. Bibi, H. R. Kaback, and G. Leblanc. 1996. Membrane topology of the melibiose permease of Escherichia coli studied by melB-phoA fusion analysis. Biochemistry 35:4161-4168. [DOI] [PubMed] [Google Scholar]
- 28.Rather, P. N., H. Munayyer, P. A. Mann, R. S. Hare, G. H. Miller, and K. J. Shaw. 1992. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase Ib and IIa proteins. J. Bacteriol. 174:3196-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 30.Sarno, R., G. McGillivary, D. J. Sherratt, L. Actis, and M. E. Tolmasky. 2002. Complete sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 46:3422-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shmara, A., N. Weinsetel, K. J. Dery, R. Chavideh, and M. E. Tolmasky. 2001. Systematic analysis of a conserved region of the aminoglycoside 6′-N-acetyltransferase type Ib. Antimicrob. Agents Chemother. 45:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strom, M. S., and S. Lory. 1987. Mapping of export signals of Pseudomonas aeruginosa pilin with alkaline phosphatase fusions. J. Bacteriol. 169:3181-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Studier, F., A. Rosenberg, J. Dunn, and J. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 35.Summers, D. K., and D. J. Sherratt. 1988. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 7:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, P. R., J. Schwartzenhauer, D. W. Hughes, A. M. Berghuis, and G. D. Wright. 1999. The COOH terminus of aminoglycoside phosphotransferase (3′)-IIIa is critical for antibiotic recognition and resistance. J. Biol. Chem. 274:30697-30706. [DOI] [PubMed] [Google Scholar]
- 39.Tolmasky, M. E. 1990. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid 24:218-226. [DOI] [PubMed] [Google Scholar]
- 40.Tolmasky, M. E., R. M. Chamorro, J. H. Crosa, and P. M. Marini. 1988. Transposon-mediated amikacin resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 32:1416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tolmasky, M. E., and J. H. Crosa. 1993. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid 29:31-40. [DOI] [PubMed] [Google Scholar]
- 42.Tolmasky, M. E., and J. H. Crosa. 1987. Tn1331, a novel multiresistance transposon encoding resistance to amikacin and ampicillin in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 31:1955-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran van Nhieu, G., and E. Collatz. 1987. Primary structure of an aminoglycoside 6′-N-acetyltransferase AAC(6′)-4, fused in vivo with the signal peptide of the Tn3-encoded beta-lactamase. J. Bacteriol. 169:5708-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vazquez-Laslop, N., H. Lee, R. Hu, and A. A. Neyfakh. 2001. Molecular sieve mechanism of selective release of cytoplasmic proteins by osmotically shocked Escherichia coli. J. Bacteriol. 183:2399-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vetting, M. W., S. S. Hegde, F. Javid-Majd, J. S. Blanchard, and S. L. Roderick. 2002. Aminoglycoside 2′-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates. Nat. Struct. Biol. 9:653-658. [DOI] [PubMed] [Google Scholar]
- 46.Vliegenthart, J. S., P. A. Ketelaar-van Gaalen, J. Eelhart, and J. A. van de Klundert. 1991. Localisation of the aminoglycoside-(3)-N-acetyltransferase isoenzyme II in Escherichia coli. FEMS Microbiol. Lett. 65:101-105. [DOI] [PubMed] [Google Scholar]
- 47.Witholt, B., M. Boekhout, M. Brock, J. Kingma, H. V. Heerikhuizen, and L. D. Leij. 1976. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal. Biochem. 74:160-170. [DOI] [PubMed] [Google Scholar]
- 48.Wolf, E., A. Vassilev, Y. Makino, A. Sali, Y. Nakatani, and S. K. Burley. 1998. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell 94:439-449. [DOI] [PubMed] [Google Scholar]
- 49.Woods, G., and J. Washington. 1995. Antibacterial susceptibility tests: dilution and disk diffusion methods, p. 1327-1341. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. ASM Press, Washington, D.C.
- 50.Wright, G. D. 1999. Aminoglycoside-modifying enzymes. Curr. Opin. Microbiol. 2:499-503. [DOI] [PubMed] [Google Scholar]
- 51.Wybenga-Groot, L. E., K. Draker, G. D. Wright, and A. M. Berghuis. 1999. Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Struct. Fold Des. 7:497-507. [DOI] [PubMed] [Google Scholar]