Abstract
The antibiotic tiamulin targets the 50S subunit of the bacterial ribosome and interacts at the peptidyl transferase center. Tiamulin-resistant Escherichia coli mutants were isolated in order to elucidate mechanisms of resistance to the drug. No mutations in the rRNA were selected as resistance determinants using a strain expressing only a plasmid-encoded rRNA operon. Selection in a strain with all seven chromosomal rRNA operons yielded a mutant with an A445G mutation in the gene coding for ribosomal protein L3, resulting in an Asn149Asp alteration. Complementation experiments and sequencing of transductants demonstrate that the mutation is responsible for the resistance phenotype. Chemical footprinting experiments show a reduced binding of tiamulin to mutant ribosomes. It is inferred that the L3 mutation, which points into the peptidyl transferase cleft, causes tiamulin resistance by alteration of the drug-binding site. This is the first report of a mechanism of resistance to tiamulin unveiled in molecular detail.
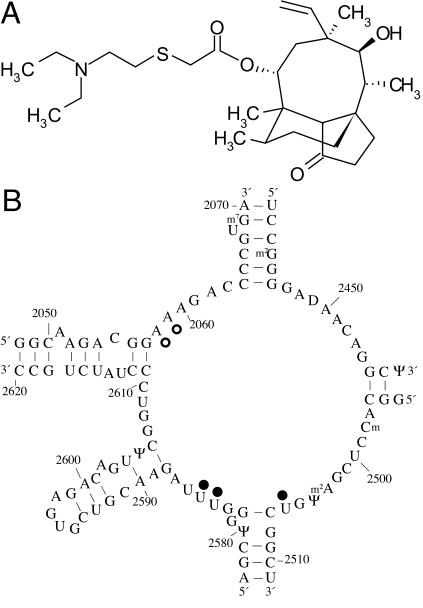
The antibiotic tiamulin (Fig. 1A) is a semisynthetic derivative of the tricyclic diterpenoid compound pleuromutilin, which was isolated from the basidiomycete Pleurotus mutilus (now called Clitopilus scyphoides) (11). Pleuromutilins inhibit protein synthesis by specifically targeting the large subunit of the bacterial ribosome. A recent study utilized chemical footprinting to reveal details of the binding of tiamulin and the related drug valnemulin to 23S rRNA (18). Nucleotides A2058 and -9, U2506, and U2584 and -5 exhibit altered reactivities in the presence of the antibiotics, indicating that they bind at the ribosomal peptidyl transferase center (Fig. 1B). Competitive footprinting experiments with macrolide antibiotics show that tiamulin and valnemulin can bind to the ribosome concurrently with erythromycin but compete with carbomycin, a peptidyl transferase inhibitor (18). These results are consistent with the inhibition of peptide bond formation by pleuromutilins observed in in vitro protein synthesis assays (7, 18). Together with earlier work indicating that the drugs are displaced by puromycin and chloramphenicol (8), the accumulated evidence suggests that the pleuromutilins bind at the ribosomal A site of the peptidyl transferase center.
FIG. 1.
(A) Chemical structure of tiamulin; (B) chemical footprint of tiamulin on 23S rRNA indicated on the secondary structure of domain V of E. coli 23S rRNA. Nucleotides with altered reactivities in the presence of tiamulin are indicated with filled circles (protections) or open circles (enhancements).
Tiamulin is used exclusively in veterinary medicine to treat swine dysentery and respiratory diseases in pigs and poultry. In some countries, only a few antibiotics are approved for use in pigs, and some of the drugs have limited effectiveness due to increased bacterial resistance. Therefore, tiamulin is an important option for the eradication of Brachyspira hyodysenteriae, a causative agent of swine dysentery, from infected herds. Resistance to the drugs tylosin and lincomycin in B. hyodysenteriae develops rapidly and is caused by a single point mutation in 23S rRNA at nucleotide A2058 (9). In contrast, tiamulin resistance develops relatively slowly and in a stepwise fashion in vitro (10). However, there has been an alarming increase in the number of isolates with decreased tiamulin susceptibility and reports of tiamulin-resistant isolates from different parts of Europe. To date, the genetic basis of tiamulin resistance is unknown. Knowledge of antibiotic resistance mechanisms is important not only for the design of improved antimicrobial agents but also for guiding policies on the current set of antibiotics so they can be used most effectively.
In this study, the genetic basis of tiamulin resistance in Escherichia coli was investigated. Strains of E. coli containing a plasmid-encoded or seven chromosomal rRNA operons were used to select for tiamulin-resistant mutants. rRNA mutations were not found as resistance determinants in the isolated mutants with plasmid-encoded rRNA, although the footprinting data point to a binding site in the rRNA. Additional selections produced tiamulin-resistant mutants with various phenotypes. In one isolate we identified a point mutation in ribosomal protein L3 responsible for the tiamulin resistance phenotype. The mutation is located in a nonglobular domain of L3 that extends close to the site of peptide bond formation. We conclude that the mutation in ribosomal protein L3 confers the observed tiamulin resistance phenotype by affecting the binding and inhibitory action of tiamulin at the peptidyl transferase center.
MATERIALS AND METHODS
E. coli strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
E. coli strains and plasmids
| Strain or plasmid | Relevant genotype and characteristics | Reference or source |
|---|---|---|
| Strains | ||
| TA542 | Δ7 (ΔEBHGcADcCc recA56/pTRNA66, pHK-rrnC+) | 1 |
| CN2476 | [CN1709] ade::cam CN1709: F−ara Δ(codB-lac)3 thi | 16, 17 |
| JB5 | [CN2476] rplC | This study |
| CAG5051-55a | nadA57::Tn10 btuB3191::Tn10 zbe-280::Tn10 trpB83::Tn10 zed-3069::Tn10 | 19 |
| CAG8209a | zgh-3075::Tn10 | 19 |
| CAG8160a | thi-39::Tn10 | 19 |
| CAG12152a | zgi-3075::Tn10 | 13 |
| CAG12072a | zha-203::Tn10 | 13 |
| CAG12075a | zhe-3083::Tn10 | 13 |
| PG201a | kefB20::Tn10 | I. Booth via EGSCb |
| Plasmids | ||
| pFK1 | rrnB operon inserted into Tetr gene of pBR322; Ampr | 4 |
| pCN102A | lac promoter and polylinker inserted downstream of the fourth codon of the lacZ gene; Ampr | 15 |
Only the position of Tn10 is given.
EGSC, E. coli Genetic Stock Center, Yale University, New Haven, Conn.
Isolation and characterization of tiamulin-resistant mutants in a strain with a plasmid-encoded rRNA operon.
Antibiotic sensitivity testing showed that the growth of E. coli strain TA542 is completely inhibited at 60 μg of tiamulin per ml. Twenty-two spontaneous mutants with decreased tiamulin sensitivity were isolated by growing strain TA542 in the presence of 60 to 100 μg of tiamulin per ml. Plasmid DNA from the mutant strains was isolated and transformed into a strain that had not been exposed to tiamulin, where pHK-rrnC+ in TA542 was replaced with pFK1 (4). Tiamulin susceptibilities of strains containing the wild-type TA542 strain background and pHK-rrnC+ from each of the mutant strains were determined by testing for growth on NZY plates containing tiamulin. In all cases, the tiamulin susceptibility of a strain containing pHK-rrnC+ isolated from a mutant was indistinguishable from that of TA542.
Isolation and genetic mapping of tiamulin-resistant mutants in a strain with seven chromosomal rRNA operons.
Seventeen spontaneous mutants with decreased tiamulin sensitivity were isolated by growing strain CN2476 on agar plates containing 150 or 200 μg of tiamulin per ml. The position of the mutation in one mutant strain, named JB5, was mapped by using a general strategy described previously (19). Two sets of strains marked with tetracycline resistance were used in the mapping procedure (Table 1), including a set of Hfr strains (CAG5051 to CAG5055, CAG8160, and CAG8209) and a set of strains for P1 transductions (CAG12072, CAG12075, CAG12152, and PG201). Hfr matings and generalized transductions with lysates of bacteriophage P1vir were performed essentially as described previously (12).
Construction of L3-lacZ expression vectors and complementation experiments.
A portion of the E. coli S10 operon from CN2476 and JB5 strains including the last 16 codons of the S10 gene, the entire L3 gene, and the first 16 codons of L4 gene was amplified by PCR and inserted between the BamHI and HindIII sites of the pCN102A vector (15). The cloned inserts were sequenced to verify their identities. The resulting L3-lacZ expression vectors (named pCN102A-wt L3 and pCN102A-mt L3, denoting plasmids containing the wild-type and mutant L3 sequences, respectively), as well as the parent vector, pCN102A, were transformed individually into CN2476 and JB5 strains. For MIC determination, overnight liquid cultures were diluted and 102 to 103 cells were spread on NZY agar plates containing tiamulin and ampicillin. MICs are the results of at least three independent experiments and are expressed as ranges, where the lower value represents the highest antibiotic concentration at which single colonies (>0.25 mm in diameter) were formed and the higher value is the lowest antibiotic concentration at which single colonies (>0.25 mm in diameter) were not formed on NZY agar plates after 48 h of growth at 37°C.
Footprinting experiments.
Ribosome isolation, chemical modification, and primer extension procedures were carried out essentially as described elsewhere (18). Primer 2654 (5′-TCCGGTCCTCTCGTACT-3′), complementary to nucleotides 2654 to 2670 of 23S rRNA, was used in the extension reaction. The cDNA products of the primer extension reactions were separated on 8% polyacrylamide-7 M urea sequencing gels.
RESULTS AND DISCUSSION
Search for tiamulin-resistant mutants with alterations in the rRNA.
Several lines of evidence indicate that tiamulin binds in a cavity lined with rRNA nucleotides at the peptidyl transferase center. For many of the peptidyl transferase antibiotics, single nucleotide alterations in the peptidyl transferase loop have been found to confer antibiotic resistance. In order to select for mutations in rRNA that could lead to tiamulin resistance, an E. coli strain with a plasmid-borne rRNA operon and seven inactivated chromosomal rRNA operons was used (Table 1) (1). Plasmid DNA from the mutant strains was isolated and transformed individually into the parent strain, where pHK-rrnC+ was replaced with pFK1 (see Materials and Methods and Table 1). The isolated plasmid did not confer a tiamulin-resistant phenotype for any of the selected mutants. Thus, the tiamulin resistance determinants obtained do not reside on the plasmid and in these cases are therefore not due to mutations in rRNA. The reason that no tiamulin-resistant rRNA mutants were selected could be that the antibiotic binding pocket is lined with essential nucleotides that are necessary for cell growth. Another explanation is that such a plasmid-borne mutation is not viable or frequent enough to be manifested as a resistant phenotype under the investigated conditions. Preliminary investigations (data not shown) indicate that the cause of decreased sensitivity to tiamulin may be nonribosomal in some cases, although these strains have not been characterized further.
Isolation and genetic mapping of a tiamulin-resistant E. coli mutant.
A second selection was carried out with an E. coli strain containing all seven chromosomal rRNA operons and a gene conferring chloramphenicol resistance to facilitate genetic mapping (Table 1) (15, 16). Spontaneous mutants were isolated after growth on agar plates containing 150 or 200 μg of tiamulin per ml. One strain, JB5, was selected for further characterization due to its relatively rapid growth in the presence of tiamulin. Strain JB5 exhibits at least a fourfold decrease in sensitivity to tiamulin compared to the parental strain (Table 2). A two-step genetic mapping strategy, involving mating with a set of Hfr strains followed by P1 transductions, was used to identify the genetic determinant of the phenotype (19). The Hfr mating experiment enabled localization of the mutation to the region bounded by min 69 to 83 on the E. coli chromosome. A series of P1 transductions using strains marked with tetracycline resistance in the relevant region indicated that the mutation is contained within min 73.9 to 74.6, a region of the genome that encodes approximately half of the ribosomal proteins. Sequencing of selected genes in the S10 operon including rplC, rplD, and rplV showed that JB5 contained a point mutation in rplC, the gene encoding ribosomal protein L3. The mutation, A445G, results in an Asn149Asp amino acid change. This result is consistent with an earlier report that associated tiamulin resistance with alterations in ribosomal proteins L3 and L4, although the molecular basis of this connection was never characterized (3).
TABLE 2.
Susceptibilities of strains to tiamulin
| Strain | Plasmida | Tiamulin MIC (μg/ml) |
|---|---|---|
| CN2476 (parent) | None | 100-125 |
| CN102A | 100-125 | |
| CN102A-wt L3 | 100-125 | |
| CN102A-mt L3 | 150-175 | |
| JB5 (L3 mutant) | None | >400 |
| CN102A | >400 | |
| CN102A-wt L3 | 200-250 | |
| CN102A-mt L3 | 300-400 |
wt, wild type; mt, mutant.
The Asn149Asp mutation in ribosomal protein L3 confers the tiamulin resistance phenotype.
Two approaches were used to obtain genetic evidence that the Asn149Asp mutation is responsible for the tiamulin resistance phenotype. First, a group of 30 transductants, composed of 15 tiamulin-resistant and 15 tiamulin-sensitive colonies, was sequenced in the region of the mutation. In every case, there was a 1:1 correspondence between the presence of the mutation and the tiamulin resistance phenotype. Second, complementation experiments were used to determine the effect of overexpressing wild-type and mutant copies of rplC on the tiamulin resistance phenotype. Two L3-lacZ expression vectors were constructed by inserting a fragment of the S10 operon flanking rplC containing either the wild-type or the mutant rplC sequence downstream of the fourth codon of the lacZ gene carried on a plasmid. The vectors were transformed into parent and mutant strains, and their subsequent tiamulin susceptibilities were determined. The MIC for the wild-type strain increased upon plasmid-encoded expression of the mutated rplC sequence together with chromosome-encoded expression of wild-type rplC, whereas plasmid-encoded expression of the wild-type rplC sequence did not measurably affect the MIC (Table 2). The marginal increase in the MIC may reflect reduced expression of the mutated rplC sequence, lower incorporation of the mutant L3 protein into ribosomes, or poor usage of mutated ribosomes when in competition with those containing the wild-type genome-encoded L3 protein. Similarly, the MIC for the mutant strain was only slightly affected by plasmid-encoded expression of the mutated rplC sequence together with chromosome-encoded expression of mutant rplC but decreased significantly upon plasmid-encoded expression of the wild-type rplC sequence (Table 2). In addition, the parent vector did not measurably affect the MIC in either the wild-type or mutant strain (Table 2). The data show that the degree of tiamulin sensitivity decreases in the wild-type strain by expression of the mutated sequence and increases in the mutant strain by expression of the wild-type L3 sequence. We conclude that the mutation yielding an Asn149Asp mutation in ribosomal protein L3 is responsible for the tiamulin resistance phenotype.
The mutation is located in a region of L3 close to the peptidyl transferase center.
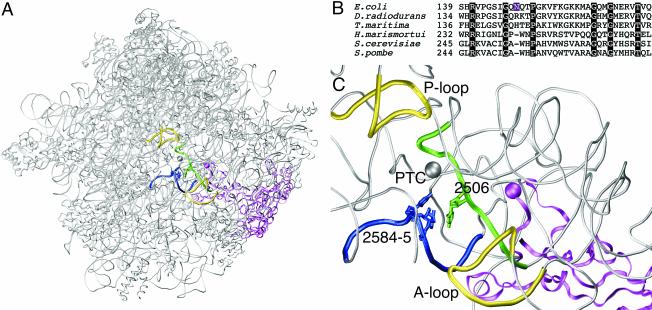
Like many ribosomal proteins, L3 has a globular domain on the ribosome surface plus an extended domain that stretches into the ribosome interior (Fig. 2A). L3 is one of four ribosomal proteins that come closest to the site of peptide bond formation through its extended domain (14). An alignment of selected L3 sequences was carried out to determine whether the mutation occurred at a conserved position in the sequence (Fig. 2B). Although Asn149 itself is not conserved, it lies within a conserved region of the protein with enough amino acid identity to obtain a good alignment between sequences representing the three domains of life. The L3 sequences of Haloarcula marismortui and Deinococcus radiodurans were included to enable localization of the mutation in the three dimensional structures of the large ribosomal subunit solved recently through X-ray crystallography (2, 6). Only the H. marismortui structure provides the resolution needed to position the amino acid side chains. The positions in H. marismortui and D. radiodurans that are equivalent to Asn149 in E. coli are Trp242 and Arg144, respectively. Trp242 is located at the tip of the extended domain and represents the position on L3 that comes closest to the catalytic center (Fig. 2C). The shortest distance between Trp242 and U2541 (U2506 in E. coli numbering), one of the nucleotides in the tiamulin footprint, is about 7 Å. Although amino acid side chains are not included in the D. radiodurans 50S structure, the backbone position of Arg144 is also the closest point on L3 to the peptidyl transferase site. The distance between the α-carbon of Arg144 and the phosphorus atom of nucleotide U2485 (U2506 in E. coli numbering) is approximately 10 Å. Interestingly, alteration of ribosomal protein L3 at the relevant amino acid also affects the action of another class of antibiotics, called the trichothecenes, that specifically target eukaryotic cells. The mutation Trp255Cys at the equivalent amino acid position causes resistance to the antibiotic trichodermin in Saccharomyces cerevisiae (5).
FIG. 2.
(A) The position of ribosomal protein L3 in the large ribosomal subunit. The subunit is shown in the crown view, where the peptidyl transferase center is in the middle of the figure. Ribosomal protein L3 is shown as a magenta ribbon, and the position of the mutation is marked with a sphere of the same color. The other highlighted items and their coloring are as described for panel C. Other ribosomal proteins and rRNA in the large ribosomal subunit are depicted as gray tubes. The coordinates of the H. marismortui 50S subunit are used (PDB accession number 1JJ2). (B) Sequence comparison of various L3 ribosomal proteins in the regions flanking the E. coli Asn149Asp mutation. The other L3 sequences are from the bacteria D. radiodurans and Thermotoga maritima, the archaeon H. marismortui, and the eukaryotes S. cerevisiae and Schizosaccharomyces pombe. The position of the mutation in E. coli is highlighted in magenta, and positions of amino acid identity are highlighted in black. (C) Close-up view of the location of the L3 mutation relative to rRNA nucleotides at the peptidyl transferase center. Ribosomal protein L3 is depicted as in panel A. The nucleotides in the tiamulin footprint are shown in green (U2506) and blue (U2584 and U2585), and the respective regions of 23S rRNA flanking these nucleotides are highlighted in the same color (nucleotides are labeled using E. coli numbering). The peptidyl transferase center (PTC) is indicated by a gray sphere centered on the N-3 atom of A2486 (A2451 in E. coli numbering). The A and P loops of 23S rRNA, bound transiently by tRNAs in the A and P sites, respectively, are depicted as yellow tubes. The gray tubes represent parts of domain V of H. marismortui 23S rRNA.
Ribosomes containing the L3 mutation exhibit reduced tiamulin binding.
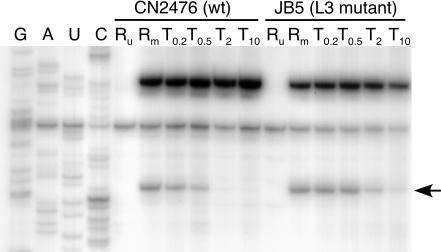
Chemical footprinting was used to examine tiamulin binding to wild-type versus L3 mutated ribosomes. Ribosome-tiamulin complexes were probed with CMCT, which modifies the N-3 of uridine residues. Primer extension with reverse transcriptase was then used to identify alterations in protection of U2506 induced by tiamulin binding to the wild-type and mutant ribosomes at drug concentrations ranging between 0.1 and 10 μM. In the wild-type ribosomes, complete protection at U2506 was observed at tiamulin concentrations above 0.5 μM (Fig. 3). In contrast, complete protection at U2506 was not observed in the mutant ribosomes even at a tiamulin concentration of 10 μM. These results reveal that tiamulin binding to mutant ribosomes is significantly reduced relative to binding to wild-type ribosomes.
FIG. 3.
Gel autoradiogram showing the protection effect at nucleotide U2506 in E. coli 23S rRNA caused by tiamulin binding to CN2476 (wild-type [wt]) and JB5 (L3 mutant) ribosomes. Dideoxy sequencing lanes are indicated by G, A, U, and C. Lanes are labeled to denote reactions with chemically unmodified 70S ribosomes in the absence of tiamulin (Ru), 70S ribosomes modified with CMCT in the absence of tiamulin (Rm), or 70S ribosomes modified in the presence of tiamulin (T0.2, T0.5, T2 or T10, where the subscript indicates the drug concentration [micromolar]). The arrow shows the altered reactivity at position U2506.
Concluding remarks.
It is well known that single nucleotide or amino acid changes in ribosomal components can lead to antibiotic resistance. In this investigation, a point mutation in the gene encoding ribosomal protein L3 was mapped in a tiamulin-resistant mutant strain and shown to be a tiamulin resistance determinant in E. coli. The mutation produces an Asn-to-Asp alteration at position 149 of ribosomal protein L3 that is in the vicinity of the peptidyl transferase site. We conclude that the mutation perturbs the drug-binding site at the peptidyl transferase center. As the mutated position seems accessible from the peptidyl transferase center according to the X-ray structures of the large ribosomal subunit, we believe it is a direct perturbation. The mutation could function by altering the tiamulin binding site either directly, by eliminating a specific interaction with tiamulin, or indirectly, by influencing the RNA structure at the peptidyl transferase center. This is the first report of a tiamulin resistance mechanism that has been characterized on a molecular level. In addition, it is the first account of a ribosomal protein L3 mutant in bacteria that confers antibiotic resistance. It will be interesting to learn if clinical isolates from the veterinary field also contain mutations in ribosomal protein L3.
Acknowledgments
We thank Novartis for furnishing tiamulin and C. Squires for strain TA542. C. Petersen is thanked for generously providing strains, plasmid CN102A, and helpful advice. M. Karlsson is thanked for helpful discussions and information about tiamulin usage.
B.V. was supported by the Nucleic Acid Center funded by the Danish National Research Foundation. K.S.L. was supported by a fellowship from Alfred Benzons Fond and a grant from the European Commission's 5th Framework Program (grant QLK2-CT-2002-00892).
REFERENCES
- 1.Asai, T., D. Zaporojets, C. Squires, and C. L. Squires. 1999. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. USA 96:1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ban, N., P. Nissen, J. Hansen, P. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 3.Böck, A., F. Turnowsky, and G. Högenauer. 1982. Tiamulin resistance mutations in Escherichia coli. J. Bacteriol. 151:1253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douthwaite, S. R., T. Powers, J. Y. Lee, and H. F. Noller. 1989. Defining the structural requirements for a helix in 23 S ribosomal RNA that confers erythromycin resistance. J. Mol. Biol. 209:655-665. [DOI] [PubMed] [Google Scholar]
- 5.Fried, H. M., and J. R. Warner. 1981. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proc. Natl. Acad. Sci. USA 78:238-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679-688. [DOI] [PubMed] [Google Scholar]
- 7.Hodgin, L. A., and G. Högenauer. 1974. The mode of action of pleuromutilin derivatives. Effect on cell-free polypeptide synthesis. Eur. J. Biochem. 47:527-533. [DOI] [PubMed] [Google Scholar]
- 8.Högenauer, G. 1975. The mode of action of pleuromutilin derivatives. Location and properties of the pleuromutilin binding site on Escherichia coli ribosomes. Eur. J. Biochem. 52:93-98. [DOI] [PubMed] [Google Scholar]
- 9.Karlsson, M. A., C. Fellström, M. U. K. Heldtander, K.-E. Johansson, and A. Franklin. 1999. Genetic basis of macrolide and lincosamide resistance in Brachyspira (Serpulina) hyodysenteriae. Vet. Microbiol. 70:225-238. [DOI] [PubMed] [Google Scholar]
- 10.Karlsson, M., A. Gunnarsson, and A. Franklin. 2001. Susceptibility to pleuromutilins in Brachyspira (Serpulina) hyodysenteriae. Anim. Health Res. Rev. 2:59-65. [PubMed] [Google Scholar]
- 11.Kavanagh, F., A. Hervey, and W. J. Robbins. 1951. Antibiotic substances from basidiomycetes. VIII. Pleurotus mutilus (Fr.) Sacc. & Pleurotus passeckerianus Pilat. Proc. Natl. Acad. Sci. USA 37:570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Nichols, B. P., O. Shafiq, and V. Meiners. 1998. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180:6408-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 15.Petersen, C. 1987. The functional stability of the lacZ transcript is sensitive towards sequence alterations immediately downstream of the ribosome binding site. Mol. Gen. Genet. 209:179-187. [DOI] [PubMed] [Google Scholar]
- 16.Petersen, C., and L. B. Møller. 2000. Invariance of the nucleoside triphosphate pools of Escherichia coli with growth rate. J. Biol. Chem. 275:3931-3935. [DOI] [PubMed] [Google Scholar]
- 17.Petersen, C., L. B. Møller, and P. Valentin-Hansen. 2002. The cryptic adenine deaminase gene of Escherichia coli. J. Biol. Chem. 277:31373-31380. [DOI] [PubMed] [Google Scholar]
- 18.Poulsen, S. M., M. Karlsson, L. B. Johansson, and B. Vester. 2001. The pleuromutilin drugs tiamulin and valnemulin bind to the RNA at the peptidyl transferase centre on the ribosome. Mol. Microbiol. 41:1091-1099. [DOI] [PubMed] [Google Scholar]
- 19.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]