Abstract
The tuberactinomycin antibiotics are essential components in the drug arsenal against Mycobacterium tuberculosis infections and are specifically used for the treatment of multidrug-resistant tuberculosis. These antibiotics are also being investigated for their targeting of the catalytic RNAs involved in viral replication and for the treatment of bacterial infections caused by methicillin-resistant Staphylococcus aureus strains and vancomycin-resistant enterococci. We report on the isolation, sequencing, and annotation of the biosynthetic gene cluster for one member of this antibiotic family, viomycin, from Streptomyces sp. strain ATCC 11861. This is the first gene cluster for a member of the tuberactinomycin family of antibiotics sequenced, and the information gained can be extrapolated to all members of this family. The gene cluster covers 36.3 kb of DNA and encodes 20 open reading frames that we propose are involved in the biosynthesis, regulation, export, and activation of viomycin, in addition to self-resistance to the antibiotic. These results enable us to predict the metabolic logic of tuberactinomycin production and begin steps toward the combinatorial biosynthesis of these antibiotics to complement existing chemical modification techniques to produce novel tuberactinomycin derivatives.
It was recently estimated that between the years 1998 and 2030 there will be 225 million new cases of tuberculosis (TB) and 79 million TB-related deaths (40). These numbers are astonishing when one considers that treatments for this disease, in the forms of vaccines or chemotherapy, have been available for more than 50 years (29). Mycobacterium tuberculosis, the causative agent of TB, is notoriously slowly growing and during infection can persist in a latent form in many individuals. These attributes contribute to the reasons why typical chemotherapy regimens for TB last 6 to 9 months (6) and why TB is so persistent. This prolonged treatment presents significant hurdles in the development of new antibiotics and in retaining the efficacies of the antibiotics used at present. Side effects and toxicity from a particular compound can be magnified when a patient takes a drug for this length of time, and there are increased incidences of poor adherence to the chemotherapy regimen by unmonitored patients, resulting in the development of multidrug-resistant (MDR) TB infections. These facts, together with alarming interactions between human immunodeficiency virus and TB infections that can result in increased numbers of infected individuals and MDR TB (30), make it of paramount importance to develop new chemotherapy agents or introduce modifications to the agents available at present to reduce their toxicities and increase their activities against MDR TB.
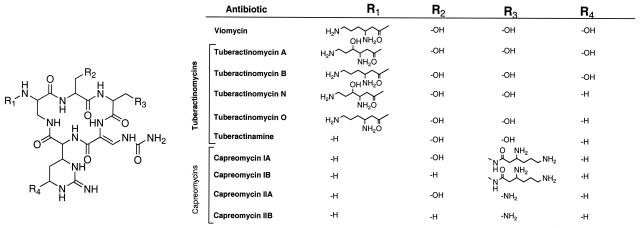
The tuberactinomycins (TUBs; this abbreviation refers to the antibiotic family as a whole) (Fig. 1) are used specifically for the treatment of MDR TB (14). The importance of TUBs is reflected by some members being included on the World Health Organization's Model List of Essential Medicines (57). In addition to their historical use for the treatment of TB, the TUBs have recently become of interest for their use for the treatment of other bacterial infections, such as those caused by vancomycin-resistant enterococci and methicillin-resistant Staphylococcus aureus (15, 32, 33), and in the targeting of catalytic RNAs to disrupt viral replication (25, 45). In these cases, the TUBs are considered encouraging lead compounds for the development of more potent drugs.
FIG. 1.
Chemical structures of the TUB family of antibiotics. The numbers within the cyclic pentapeptide core identify the residue numbers noted in the text. The figure is as presented elsewhere (55), with modifications.
The TUBs are peptide antibiotics containing nonproteinogenic amino acids; as a result, it is anticipated that these molecules are biosynthesized via a nonribosomal peptide synthetase (NRPS) mechanism. The unusual amino acids synthesized and condensed to produce these antibiotics are 2,3-diaminopropionate, β-ureidodehydroalanine, β-lysine, and l-capreomycidine, with the last two amino acids being hydroxylated at the γ carbon and the C-6 carbon, respectively, in certain members of the TUB family.
To understand how TUB antibiotics are biosynthesized, we have isolated, sequenced, and annotated the viomycin biosynthetic gene cluster from Streptomyces sp. strain ATCC 11861. Genetic inactivation of the first gene in the biosynthetic gene cluster abolishes viomycin production, confirming that this gene cluster is involved in viomycin biosynthesis. The gene cluster consists of 36.3 kb of DNA encoding 20 open reading frames (ORFs) involved in the biosynthesis, regulation, export, and activation of the antibiotic, in addition to the viomycin resistance gene. From this information, hypotheses on how all of the necessary nonproteinogenic amino acids are biosynthesized, incorporated into the hexapeptide, and modified were developed. This information sets the stage for enhancing our ability to generate new structural derivatives of the TUBs, using combinatorial biosynthesis, for the treatment of MDR TB, as well as other bacterial and viral infections.
MATERIALS AND METHODS
Bacterial strains and growth media.
Streptomyces sp. strain ATCC 11861 was obtained from the American Type Culture Collection and was grown on ISP medium 2 (0770; Difco). The strain was grown in Bacto Tryptic Soy Broth (TSB) to obtain mycelia for chromosomal DNA isolation. For the production and purification of viomycin, mycelia grown in TSB were used to inoculate 100 ml of viomycin production medium (44). Mannitol soy flour agar was used for conjugations (27).
Escherichia coli strains were grown in Luria-Bertani (LB) medium or on LB agar supplemented with the appropriate antibiotic, as indicated. When cosmid-containing strains were grown in microtiter plates, the strains were grown in freezing medium (56) supplemented with kanamycin (50 μg/ml). The E. coli strains used were DH5α, XL-1 Blue MR (Stratagene), HB101/pRK2013 (18) (from M. Rondon, University of Wisconsin—Madison), and ET12567 (34) (from C. Khosla, Stanford University).
Genomic DNA isolation and cosmid library construction.
A total of 3.0 g (wet weight) of Streptomyces sp. strain ATCC 11861 mycelia was used for genomic DNA isolation by a previously described protocol (43). Genomic DNA was partially digested with Sau3AI to give 30- to 50-kb fragments that were subsequently ligated into the BamHI site of SuperCos1 (Stratagene), prepared according to the instructions of the manufacturer. The DNA was then packaged into lambda phage by using the Gigapack III XL Packaging Extract kit (Stratagene), and the cosmid was used to infect E. coli XL-1 Blue MR, according to the instructions of the manufacturer. A total of 1,248 cosmid-containing clones were isolated and frozen at −80°C individually in microtiter dish wells as well as in pools of 8 clones consisting of 25 μl from each member of a microtiter dish column. Thus, the 1,248 individual cosmid-containing clones were also represented in 156 cosmid-containing pools.
Screening of the cosmid library.
The cosmid library was first screened by PCR amplification for those cosmids that contained vph, the viomycin resistance gene (4). The following primers were used: primer Vph/FEco (5′-AGAAGTGGAGAATTCGCCCACCATGAG-3′) and primer Vph/REco (5′-CCTTCAGAATTCCTGTCACGCTGCCCG-3′). Boiled cells of each cosmid pool were used as a source of template DNA for PCR amplification. Individual members of each vph-positive cosmid pool were subsequently screened by PCR amplification to identify the specific cosmid containing vph. Cosmid pVIO-P2C3RG was identified in this manner.
Two primers based on the putative viomycin biosynthetic gene vioG were designed from the pVIO-P2C3RG sequence, primer vio-P2-5p (5′-GGGGAGACGTACTTCTTCCA-3′) and primer vio-P2-3p (5′-GGCGAGTTCACGGGAGATA-3′). These primers were used to screen the library a second time by PCR amplification to identify cosmids containing vioG. The vioG-positive cosmids were then screened by PCR amplification for the absence of vph. A vioG-positive but vph-negative cosmid, pVIO-P8C8RH, was thus isolated.
Sequencing and annotation of the viomycin biosynthetic gene cluster.
Fragments of 2 to 3 kb from cosmids pVIO-P2C3RG and pVIO-P8C8RH were subcloned into pSMARTLCKan by Lucigen Corp. (Middleton, Wis.). Subclones were submitted to the Genome Center Sequencing Facility at University of Wisconsin—Madison, where they were sequenced (sevenfold coverage, twofold minimum). Contigs were assembled by using the SeqMan program (Lasergene, Madison, Wis.). Annotation of ORFs and putative gene functions were assigned by using a combination of MapDraw (Lasergene) and the blastp, PSI-BLAST, and RPS-BLAST (National Center for Biotechnology Information) programs (2).
Insertional inactivation of vioA.
An internal fragment of vioA was introduced into the suicide vector pOJ260 (5) by PCR-based cloning. The primers used for vioA amplification by PCR were as follows: primer VioA/Pst (5′-TCACGCCGGTCGAGCAGGA-3′) and primer VioA/Eco (5′-ACGCCGTACTCGCGCAGG-3′). The PCR-amplified product was digested with PstI and EcoRI and cloned into the corresponding restriction sites of pOJ260, yielding pOJ260-vioA. This plasmid was transformed into ET12567, and the resulting strain was used for conjugation of pOJ260-vioA into Streptomyces sp. strain ATCC 11861 by a triparental mating protocol (27). The triparental mating involved ET12567/pOJ260-vioA, HB101/pRK2013, and Streptomyces sp. strain ATCC 11861.
To confirm pOJ260-vioA insertion into the chromosomal copy of vioA, chromosomal DNA was purified from the mutant strains and analyzed by PCR amplification and subsequent restriction enzyme analysis of the amplified products. The primers used for this analysis were primers VioA/Pst and VioA/Eco, which anneal to regions outside the region cloned into pOJ260, and primer FOR (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and primer REV (5′-TCACACAGGAAACAGCTATGA-3′), which anneal to regions just outside the multiple cloning site of pOJ260. During this analysis it was determined that the 5′ end of vioA in one mutant strain (strain MGT1001) had undergone a deletion of approximately 400 bp between a BglII and an NcoI restriction site within vioA (data not shown). This was not characterized further because vioA was inactivated regardless of the nature of this deletion. The two other vioA mutants isolated (strains MGT1002 and MGT1003) did not contain this deletion (data not shown).
Production and isolation of viomycin.
Cultures of 100 ml of the wild-type or mutant strains of Streptomyces sp. strain ATCC 11861 were grown in viomycin production medium at 28°C for 5 days. Mycelia were removed by centrifugation, and the resulting supernatant was used for viomycin purification by a previously published protocol (48).
HPLC of purified viomycin.
Purified viomycin samples were analyzed by high-performance liquid chromatography (HPLC; Beckman System Gold) with a Macrosphere SCX 300A 7U column (Alltech) at a flow rate of 1 ml/min. The following buffers were used: buffer A (20 mM Tris-HCl [pH 6.4]) and buffer B (20 mM Tris-HCl, 1 M sodium acetate [pH 6.4]). The separation profile was 5 min of isocratic development with 100% buffer A-0% buffer B, 15 min of a linear gradient from 100% buffer A-0% buffer B to 0% buffer A-100% buffer B, and 5 min of isocratic development with 0% buffer A-100% buffer B. Elution of viomycin was monitored at the characteristic absorbance of 268 nm. The purified viomycin had the same UV and visible light spectra and HPLC retention time as authentic viomycin and also coeluted from the high-performance liquid chromatograph with authentic antibiotic, regardless of the elution profile.
Nucleotide sequence accession number.
The completed viomycin biosynthetic gene cluster has been deposited in GenBank under accession no. AY263398.
RESULTS AND DISCUSSION
Cloning and sequencing of the viomycin biosynthetic gene cluster.
We constructed a cosmid library of the Streptomyces sp. strain ATCC 11861 genome and used PCR amplification to screen the library for cosmids containing vph, the known viomycin resistance gene (4). The resistance gene was targeted because the resistance gene for a particular antibiotic is typically encoded in the same region of the chromosome as the biosynthetic gene cluster for that antibiotic (37). Sequencing out of the resistance gene from one of the vph-positive cosmids, pVIO-P2C3RG, identified an ORF that encoded a putative lysine 2,3-aminomutase. Since viomycin contains a β-lysine moiety and lysine 2,3-aminomutases catalyze the formation of β-lysine, we hypothesize that pVIO-P2C3RG contains a portion of the viomycin biosynthetic gene cluster.
Preliminary analysis of the DNA sequence from pVIO-P2C3RG suggested that only a portion of the viomycin biosynthetic gene cluster was contained on the cosmid. We then screened the library a second time using PCR primers whose sequences were based on a putative viomycin biosynthetic gene, vioG, that was present on pVIO-P2C3RG. The vioG-positive cosmids were then screened for the absence of vph, and cosmids containing DNA that overlapped but that was not redundant with the insert in pVIO-P2C3RG were identified. From this analysis, pVIO-P8C8RH was isolated and both cosmids were completely sequenced.
Analysis of the viomycin biosynthetic gene cluster.
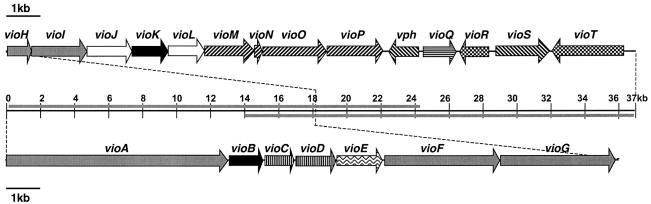
We predict that the viomycin biosynthetic gene cluster includes approximately 36.3 kb of contiguous DNA encoding 20 ORFs involved in the biosynthesis, export, regulation, and activation of the antibiotic, in addition to the previously isolated resistance gene vph (Fig. 2; Table 1). An additional ∼20 kb on either side of the predicted gene cluster was sequenced, and analysis of this DNA did not identify any genes predicted to be involved in viomycin production. Thus, we predict that vioA, vioT, and the genes between them constitute the viomycin biosynthetic gene cluster.
FIG. 2.
Schematic representation of the viomycin biosynthetic gene cluster. The center line denotes the 37 kb encoding all ORFs involved in viomycin biosynthesis, with the bars above and below the center line representing DNA present on cosmids pVIO-P8C8RH and pVIO-P2C3RG, respectively. Arrows above and below the center line identify the direction of transcription of ORFs. Coding of ORF biosynthetic function is as follows: grey, NRPS; black, l-2,3-diaminopropionate; white, l-2,3-diaminopropionate→β-ureidodehydroalanine; vertical lines, l-capreomycidine; horizontal lines, l-capreomycidine hydroxylation; right-slanted lines, β-lysine; left-slanted lines, resistance and activation; checkerboard pattern, regulation; waves, export.
TABLE 1.
Summary of genes and predicted encoded functions
| Gene | Start-stop positions | Predicted encoded functiona |
|---|---|---|
| vioA | 415-6786 | NRPS (A-PCP-C-A-PCP-C) |
| vioB | 6981-8021 | 2,3-Diaminopropionate synthase |
| vioC | 8018-9094 | l-Arg hydroxylase |
| vioD | 9091-10260 | Capreomycidine synthase |
| vioE | 10257-11600 | Permease |
| vioF | 11597-14818 | NRPS (A-PCP-C) |
| vioG | 14908-18174 | NRPS (A-PCP-C/-?) |
| vioH | 18171-18959 | Type II thioesterase |
| vioI | 18956-20608 | NRPS (PCP-C) |
| vioJ | 20605-21777 | 2,3-Diaminopropionyl α,β-desaturase |
| vioK | 21827-22909 | Ornithine cyclodeaminase |
| vioL | 22906-23832 | Carbamoyltransferase |
| vioM | 23829-25202 | NRPS (C)-β-lysine transferase |
| vioN | 25199-25390 | MbtH homolog |
| vioO | 25396-27228 | NRPS (A-PCP)-β-lysine activation |
| vioP | 27303-28640 | Lysine 2,3-aminomutase |
| vph | 29557-28676 | Viomycin phosphotransferase |
| vioQ | 29590-30621 | Capreomycidine hydroxylase |
| vioR | 31397-30660 | Transcriptional regulator |
| vioS | 31896-33110 | Viomycin-phosphate phosphatase |
| vioT | 36299-33717 | Trascriptional regulator |
Abbreviations for NRPS domains: C/, truncated condensation; ?, domain of unknown function. The other abbreviations are defined in the text.
Biosynthesis of the nonproteinogenic amino acids.
Viomycin is a 6-amino-acid peptide consisting of two l-serine residues and one residue of each of the following nonproteinogenic amino acids: l-2,3-diaminopropionate, β-ureidodehydroalanine, β-lysine, and l-tuberactidine. On the basis of the common observation that secondary metabolite biosynthetic gene clusters typically encode all the enzymes needed for the production of any precursors specific for that particular metabolite (12, 16, 53), we predict that the viomycin gene cluster should encode the enzymes needed to generate l-2,3-diaminopropionate, l-capreomycidine, and β-lysine. The conversion of l-2,3-diaminopropionate to β-ureidodehydroalanine and the conversion of l-capreomycidine to l-tuberactidine are predicted to occur after precursor incorporation into the growing peptide chain, as will be discussed below.
(i) Biosynthesis of l-2,3-diaminopropionate.
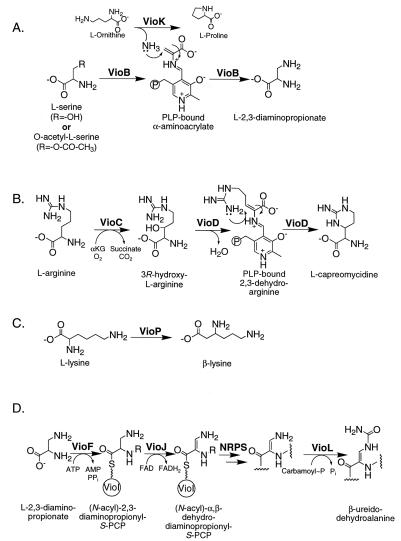
Precursor labeling studies with viomycin (9) and the capreomycins (54) have determined that l-serine is the precursor for l-2,3-diaminopropionate. Bioinformatic analysis of the viomycin biosynthetic gene cluster suggests that the conversion of l-serine to l-2,3-diaminopropionate is catalyzed by the concerted actions of VioB and VioK (Fig. 3A).
FIG. 3.
Schematic representations of the biosynthetic pathways for the four nonproteinogenic amino acids in Vio. The abbreviations are defined in the text. In panel D, R is —H or —tripeptide, and the last step in β-ureidodehydroalanine biosynthesis occurs after cyclic pentapeptide synthesis.
VioB is a homolog of cysteine synthases and serine dehydratases, enzymes that catalyze the pyridoxal phosphate (PLP)-dependent replacement and elimination, respectively, of the β substituents of their substrates (1). During catalysis, both of these enzymes form a PLP-bound α-aminoacrylate intermediate. We predict that VioB uses a similar mechanism to form a Schiff base linkage between PLP and an α-aminoacrylate intermediate (Fig. 3A). VioB then catalyzes a β-substituent replacement reaction analogous to that seen for cysteine synthases. However, while cysteine synthases use sulfur from sulfide as the nucleophile, we predict that VioB uses the nitrogen of ammonia as the nucleophile (Fig. 3A). The source of this nucleophile for viomycin biosynthesis is the ammonia liberated from l-ornithine by VioK (Fig. 3A). VioK is a homolog of ornithine cyclodeaminases, enzymes that convert l-ornithine to l-proline with the release of ammonia (13, 41, 42). Thus, VioK is predicted to function as an amidotransferase during viomycin biosynthesis since ammonia, not l-Pro, is the relevant product of the reaction.
(ii) Biosynthesis of l-capreomycidine.
Precursor labeling studies have determined that l-tuberactidine of viomycin (10) and l-capreomycidine of the capreomycins (22) are derived from l-arginine. As discussed below, we hypothesize that l-capreomycidine is incorporated into the growing peptide chain and is subsequently converted to l-tuberactidine after peptide synthesis is completed. Our prediction is that VioC and VioD convert l-arginine to L-capreomycidine (Fig. 3B).
VioC is a homolog of clavaminic acid synthases, which are nonheme iron dioxygenases involved in clavulanic acid biosynthesis (52). The first reaction catalyzed by one of the clavaminic acid synthases, CS2, is the hydroxylation of the β carbon of the arginine moiety of 5-guanidino-2-(2-oxo-azetidin-1-yl)-pentanoic acid (46). We predict that VioC catalyzes a similar reaction to generate β-hydroxyarginine (Fig. 3B). This product would then be a substrate for VioD, a homolog of PLP-dependent aromatic amino acid aminotransferases. VioD would catalyze a β-elimination reaction to generate a PLP-linked α,β-dehydroarginine that would allow intramolecular addition of the δ-guanido moiety to the β carbon, thus generating l-capreomycidine.
Gould and Minott (22) predicted the presence of the α,β-dehydroarginine intermediate on the basis of their results from feeding experiments with [2,3,3,5,5-2H5]arginine during capreomycin biosynthesis. Their prediction was based on the loss of the deuterium from C-2 and the loss of one deuterium from C-3, which would be consistent with such an intermediate. They also predicted that the conversion of l-arginine to l-capreomycidine would occur after peptide synthesis so that the α,β-dehydroarginine intermediate could be stabilized by an amide bond. We predict that l-capreomycidine is produced before peptide synthesis (Fig. 3B), with the PLP cofactor stabilizing the α,β-dehydroarginine intermediate. Further experimentation will be needed to differentiate between these hypotheses.
The two putative enzymes showing the highest amino acid identity with VioC and VioD are SttL and SttM, respectively. The genes encoding SttL and SttM are within the proposed biosynthetic gene cluster for the broad-spectrum antibiotic streptothricin (17), and l-capreomycidine is predicted to be an intermediate in the biosynthesis of this antibiotic (20, 21, 24, 38). Thus, we predict that the l-capreomycidine intermediate in streptothricin biosynthesis is generated by a mechanism analogous to that proposed for viomycin.
(iii) Biosynthesis of β-lysine.
Viomycin and streptothricin also contain β-lysine moieties. Both biosynthetic clusters encode homologs to lysine 2,3-aminomutases (VioP, viomycin; SttO, streptothricin). Frey (19) has extensively studied lysine 2,3-aminomutases and has shown that these enzymes catalyze the migration of the α-amino group of l-lysine to the β carbon. We predict that both VioP and SttO catalyze the same reaction to generate a source of β-lysine for viomycin and streptothricin, respectively (Fig. 3C).
Assembly of the cyclic pentapeptide core.
Although viomycin is a peptide consisting of 6 amino acids, we predict that the cyclic pentapeptide core of the antibiotic would be biosynthesized first, followed by acylation of residue 1 with β-lysine. This hypothesis was based on the isolation of TUBs with or without a β-lysine moiety (Fig. 1), suggesting that β-lysine addition is separate from cyclic pentapeptide synthesis. We predict that a five-module NRPS synthesizes and cyclizes the pentapeptide core of viomycin.
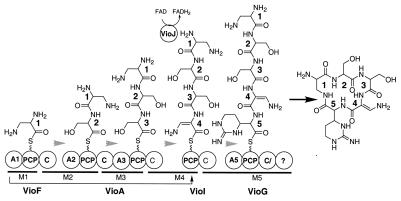
During NRPS-catalyzed peptide synthesis, the domains, modules, and subunits of these enzymes are typically aligned in a sequence that is colinear with the resulting peptide. Additionally, the organization of the NRPS subunits usually follows the order in which the corresponding genes are found on the genome (8). The viomycin NRPS does not appear to follow either of these rules. First, there are five modules for cyclic pentapeptide biosynthesis, but one of these modules lacks an adenylation (A) domain (Fig. 4). Therefore, one of the other A domains must function twice (Fig. 4). Second, it is not anticipated that the NRPS subunits function in the order in which their corresponding genes are arrayed on the chromosome (Fig. 2 and 4A).
FIG. 4.
Schematic representations of the viomycin NRPS. The domains of each subunit are shown as circles. The bars below the NRPS subunits denote specific modules, which are annotated M1 through M5. The arrow between VioF and VioI represents the in trans aminoacylation of VioI by the A1 domain of VioF. The grey arrows indicate the direction of peptide synthesis. The abbreviations for the NRPS domains are defined in footnote a of Table 1 and the text.
To determine the order in which the NRPS subunits function, we analyzed A domain specificity codes (11, 47), conserved domain sequences (28), and domain organizations. As shown in Fig. 4, we predict that VioF activates and tethers l-2,3-diaminopropionate (residue 1) to its peptidyl carrier protein (PCP) domain. Thus, the first two domains (A and PCP) of VioF would form the initiating module of the NRPS. The condensation (C) domain of VioF would then catalyze peptide bond formation between residue 1 bound to VioF and the l-serine (residue 2) bound to the first PCP domain of VioA. Thus, module 2 of the NRPS would span both VioF and VioA. The first C domain of VioA would catalyze peptide bond formation between the dipeptide (residues 1 and 2) bound to the first VioA PCP domain and l-serine (residue 3) bound to the second PCP domain of VioA. Consistent with the proposal, both A domains of VioA have specificity codes for l-serine recognition (DVYHFSLVDK and DVRHMSMVMK) (11, 47). The second C domain of VioA would then catalyze peptide bond formation between the tripeptide (residues 1, 2, and 3) on VioA and l-2,3-diaminopropionate (residue 4) bound to the PCP domain of VioI.
Two unusual aspects of the viomycin NRPS in cyclic pentapeptide synthesis need to be highlighted at this point. First, VioI lacks the necessary A domain to aminoacylate itself with l-2,3-diaminopropionate. Thus, we propose that the A domain of VioF aminoacylates VioI in trans (Fig. 4). Second, we propose that VioJ catalyzes the next step in peptide synthesis by desaturating the α,β bond of residue 4 to generate a tetrapeptide that includes the desaturated 2,3-diaminopropionate (Fig. 4). This hypothesis is based on the fact that VioJ is a homolog of acyl coenzyme A (acyl-CoA) dehydrogenases (50), but it lacks the conserved aspartate residue in acyl-CoA dehydrogenases needed for the recognition of the adenine base of CoA (3, 51). We have recently shown that acyl-CoA dehydrogenase homologs in the undecylprodigiosin and pyoluteorin pathways, which also lack this residue, catalyze α,β-desaturation of PCP-bound substrates, not their CoA derivatives (49). Thus, we propose that VioJ functions in an analogous manner and would be the first α,β-desaturase to be an integral part of a peptide synthesizing NRPS.
VioG is the best candidate for the terminal subunit since the C terminus of VioG contains a truncated C domain (immediately after the C-3 core motif), suggesting that it is inactive. This is consistent with a proposal that VioG contains the terminal module, since peptide bond formation with a downstream PCP-bound substrate is not required. This truncated C domain precedes a terminal domain of unknown function. Typically, the terminal domain of an NRPS is a thioesterase that catalyzes hydrolysis or cyclization and release from the final PCP domain (36). While a weak thioesterase motif (GSAG) could be found in this terminal domain, it is not clear whether it plays any role in peptide cyclization and release; therefore, the mechanism of pentapeptide macrocyclization remains an open question.
Modifications of the cyclic pentapeptide.
Following the formation of the cyclic pentapeptide shown in Fig. 4A, three modifications must occur: (i) carbamoylation of the β-amino group of residue 4 to form β-ureidodehydroalanine, (ii) hydroxylation of the C-6 of residue 5 to form l-tuberactidine, and (iii) acylation of the α-amino group of residue 1 with β-lysine.
VioL is predicted to catalyze the carbamoylation of residue 4, based on the amino acid similarity between VioL and ornithine carbamoyltransferases (31). The biosynthetic pathway for l-2,3-diaminopropionate conversion to β-ureidodehydroalanine is predicted to occur as shown in Fig. 3D. However, we cannot eliminate the possibility that carbamoylation occurs immediately after the desaturation of residue 4 while it is bound to VioI. Further analysis is needed to discriminate between these possibilities.
The hydroxylation of the C-6 of residue 5, which generates tuberactidine from l-capreomycidine, is likely catalyzed by VioQ, based on its similarity to phenylpentanoic acid dioxygenase and related ring-hydroxylating dioxygenases (39). We propose that the hydroxylation of residue 5 to generate the l-tuberactidine moiety occurs after peptide synthesis, based on the isolation of tuberactinomycin derivatives with and without this hydroxylation (Fig. 1), which suggests that the hydroxylation is not a prerequisite for peptide synthesis. However, as with the carbamoylation discussed above, we cannot eliminate the possibility that the hydroxylation precedes the completion of peptide synthesis.
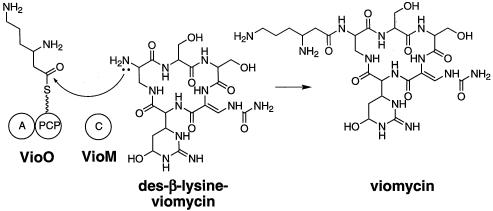
The sixth amino acid present in viomycin is β-lysine. This attachment of β-lysine to the α-amino group of residue 1 is predicted to occur by the actions of VioO (A and PCP) and VioM (C) (Fig. 5). Since these two proteins contain the three core domains of an NRPS module, these proteins can be considered a monomodular NRPS. We predict that VioO will activate and tether β-lysine to its C-terminal PCP domain. Consistent with this proposal, the A domain of VioO has the specificity code for β-lysine (DTEDVGTMVK [23]), and preliminary results with partially purified VioO suggest that it activates β-lysine (Y. Chan and M. Thomas, unpublished data).
FIG. 5.
Schematic representation of VioO- and VioM-catalyzed N-acylation of des-β-lysine-viomycin with β-lysine.
VioM, a homolog of C domains, then catalyzes amide bond formation between β-lysine bound to VioO and the soluble substrate des-β-lysine-viomycin (Fig. 5). This monomodular NRPS, therefore, does not function as a peptide synthetase per se but, rather, as an N-acyltransferase. This proposed mechanism is analogous to that for the biosynthesis of the terminal portion of the vibriobactin, whereby the acceptor site of the VibH C domain binds to a soluble substrate instead of an amino acid tethered to a PCP (26).
It is not clear what role VioN plays in viomycin biosynthesis. It is a homolog of a family of small proteins found in many NRPS systems, the standard of which is MbtH, a protein of unknown function in mycobactin biosynthesis (44). The location of vioN between vioM and vioO suggests that VioN plays some role in β-lysine addition.
Regulation, export, resistance, and activation.
Two putative transcriptional regulators are encoded in the viomycin biosynthetic gene cluster. VioR belongs to the OxyR family of transcriptional regulators, while VioT is a homolog of NysRI, a putative transcriptional regulator in the nystatin biosynthetic pathway (7). The involvement of both transcriptional regulators in viomycin biosynthesis remains to be determined.
The export of the antibiotic is likely to be catalyzed by VioE, which is a permease homolog. Our hypothesis is that viomycin is exported in its phosphorylated form. This is based on the fact that VioS is a homolog of StrK, the streptomycin phosphate phosphatase that removes the phosphate and activates streptomycin outside the cell (35). Thus, Vph, the previously identified viomycin phosphotransferase, catalyzes the phosphorylation of viomycin, which is then exported by VioE. VioS reactivates the antibiotic once it is outside the cell. Further analysis is required to determine whether this model is correct. This mechanism of resistance and export will need to be considered when the viomycin biosynthetic pathway is engineered metabolically.
Genetic evidence that the sequenced gene cluster encodes the viomycin biosynthetic enzymes.
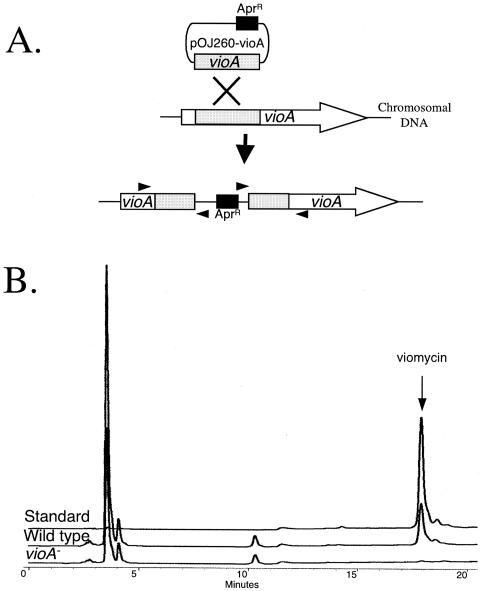
To confirm that the gene cluster that we identified is involved in viomycin biosynthesis, vioA was inactivated as shown in Fig. 6A. We chose to inactivate vioA because we propose that it encodes an essential component of the viomycin NRPS (Fig. 4) and the absence of VioA would abolish viomycin production. This result would strongly support our hypothesis that the biosynthetic gene cluster that we identified assembles viomycin. Three independently isolated vioA::pOJ260-vioA strains (strains MGT1001, MGT1002, and MGT1003) were grown under conditions optimized for viomycin production, and the antibiotic was purified from the supernatants. The purified components of the supernatant were then analyzed for the presence of viomycin by HPLC. Viomycin was not produced by any of the vioA::pOJ260-vioA strains (Fig. 6B). Therefore, we can conclude that the gene cluster sequenced and analyzed is the viomycin biosynthetic gene cluster.
FIG. 6.
(A) Schematic representation of the insertion of pOJ260-vioA into the chromosomal copy of vioA by single homologous recombination. Grey boxes, regions of identity between the cloned vioA fragment and vioA on the chromosome; arrowheads, locations of primers used to confirm the vioA::pOJ260-vioA mutations; black box and associated AprR, the apramycin resistance gene on pOJ260-vioA. Resistance to this antibiotic was used for selection of the single-crossover insertional inactivation of vioA. (B) Representative HPLC traces comparing the viomycin produced by a wild-type strain and one of the vioA-negative (vioA−) strains (strain MGT1001) of Streptomyces sp. strain ATCC 11861 to authentic viomycin (10 μg). Viomycin was not detected in vioA-negative strain MGT1002 or MGT1003 (data not shown).
Conclusions.
We have isolated, sequenced, and annotated the biosynthetic gene cluster for the antibiotic viomycin from Streptomyces sp. strain ATCC 11861. This pathway involves novel precursor biosynthetic mechanisms and atypical NRPS components in the generation of the hexapeptide antibiotic. It is anticipated that all TUB antibiotics will be biosynthesized in a manner similar to that for viomycin, with subtle changes to generate the structural diversity shown in Fig. 1.
We hypothesize that Saccharothrix mutabolis subsp. capreolus, the strain that produces the capreomycins, does not encode a VioQ homolog since the capreomycins contain l-capreomycidine, not l-tuberactidine (Fig. 1). We also predict that the tuberactinomycin producers encode an additional enzyme that catalyzes γ-hydroxylation of the β-lysine residue to generate tuberactinomycins A and N (Fig. 1).
The central pentapeptide core of the TUBs varies in two respects besides the l-capreomycidine hydroxylation discussed above. First, for viomycin and the tuberactinomycins, residue 3 of the pentapeptide core is l-serine, not l-2,3-diaminopropionate, as seen in the capreomycins (Fig. 1). This difference is one of the reasons why residue 3 of viomycin and the tuberactinomycins cannot be N-acylated with β-lysine, as seen in the capreomycins. It is anticipated that the A domain specificity code for module 3 in the capreomycin producer is altered to activate l-2,3-diaminopropionate instead of l-serine. Second, residue 2 in the capreomycins can be either l-serine or l-alanine (Fig. 1), suggesting that the A domain of module 2 of the NRPS has relaxed substrate specificity.
Finally, the capreomycins, while they contain l-2,3-diaminopropionate at residue 1 of the pentapeptide core, are N-acylated only at residue 3 (Fig. 1). It is anticipated that the VioM homolog in S. mutabolis subsp. capreolus has an altered acceptor site, which leads to β-lysine addition at the opposing side of the cyclic pentapeptide core compared to the side at which it is added in viomycin and the tuberactinomycins. This selectivity is due, in part, to the N-acylation of the β amino group of residue 3, not the α-amino group of residue 1, as seen in viomycin and the tuberactinomycins.
With the knowledge gained from the analysis of the viomycin biosynthetic gene cluster, a clear picture of how the TUB family of antibiotics is biosynthesized has been developed. Testing of the hypotheses presented here sets the stage for coordination of the metabolic engineering and chemical modification of this family of antibiotics to combat resistant bacteria, remove unwanted side effects for MDR TB treatment, and develop derivatives of these antibiotics for use in the treatment of other bacterial and viral infections.
Acknowledgments
This work was supported, in part, by Wisconsin Agricultural Experimentation Station project WIS04726 (to M.G.T.). Y.A.C. was supported by a Chemistry-Biology Interface Training Program Predoctoral Fellowship.
We thank M. R. Rondon for critical reading of the manuscript and many helpful discussions. We thank B. Shen (University of Wisconsin—Madison) for plasmid pOJ260. We thank J. Davies (University of British Columbia) for authentic viomycin.
REFERENCES
- 1.Alexander, F. W., E. Sandmeier, P. K. Mehta, and P. Christen. 1994. Evolutionary relationships among pyridoxal-5′-phosphate-dependent enzymes. Regio-specific alpha, beta and gamma families. Eur. J. Biochem. 219:953-960. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battaile, K. P., J. Molin-Case, R. Paschke, M. Wang, D. Bennett, J. Vockley, and J. J. Kim. 2002. Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases. J. Biol. Chem. 277:12200-12207. [DOI] [PubMed] [Google Scholar]
- 4.Bibb, M. J., J. M. Ward, and S. N. Cohen. 1985. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol. Gen. Genet. 199:26-36. [DOI] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Bloom, B. R., and J. D. McKinney. 1999. The death and resurrection of tuberculosis. Nat. Med. 5:872-874. [DOI] [PubMed] [Google Scholar]
- 7.Brautaset, T., O. N. Sekurova, H. Sletta, T. E. Ellingsen, A. R. StrLm, S. Valla, and S. B. Zotchev. 2000. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 7:395-403. [DOI] [PubMed] [Google Scholar]
- 8.Cane, D. E., and C. T. Walsh. 1999. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem. Biol. 6:R319-R325. [DOI] [PubMed] [Google Scholar]
- 9.Carter, J. H., II, R. H. Du Bus, J. R. Dyer, J. C. Floyd, K. C. Rice, and P. D. Shaw. 1974. Biosynthesis of viomycin. I. Origin of alpha, beta-diaminopropionic acid and serine. Biochemistry 13:1221-1227. [DOI] [PubMed] [Google Scholar]
- 10.Carter, J. H., II, R. H. Du Bus, J. R. Dyer, J. C. Floyd, K. C. Rice, and P. D. Shaw. 1974. Biosynthesis of viomycin. II. Origin of beta-lysine and viomycidine. Biochemistry 13:1227-1233. [DOI] [PubMed] [Google Scholar]
- 11.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 12.Chater, K. F., and C. J. Bruton. 1985. Resistance, regulatory and production genes for the antibiotic methylenomycin are clustered. EMBO J. 4:1893-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costilow, R. N., and L. Laycock. 1971. Ornithine cyclase (deaminating). Purification of a protein that converts ornithine to proline and definition of the optimal assay conditions. J. Biol. Chem. 246:6655-6660. [PubMed] [Google Scholar]
- 14.Croft, J., P. Chaulet, and D. Maher. 1997. Guidlines for the management of drug-resistant tuberculosis. Report WHO/TB/96.210 (rev. 1). World Health Organization, Geneva, Switzerland.
- 15.Dirlam, J. P., A. M. Belton, N. C. Birsner, R. R. Brooks, S.-P. Chang, R. Y. Chandrasekaran, J. Clancy, J. B. Cronin, B. P. Dirlam, S. M. Finegan, S. A. Froshauer, A. E. Girard, S. F. Hayashi, R. J. Howe, J. C. Kane, B. J. IKamicker, S. A. Kaufman, N. L. Kolosko, M. A. LeMay, R. G. Linde, I. I., J. P. Lyssikatos, C. P. MacLelland, T. V. Magee, M. A. Massa, S. A. Miller, M. L. Minich, D. A. Perry, J. W. Petitpas, C. P. Reese, S. B. Seibel, W.-G. Su, K. T. Sweeney, D. A. Whipple, and B. V. Yang. 1997. Cyclic homopentapeptides. 1. Analogs of tuberactinomycins and capreomycin with activity against vancomycin-resistant enterococci and Pasteurella. Bioorg. Med. Chem. Lett. 7:1139-1147. [Google Scholar]
- 16.Du, L., C. Sanchez, M. Chen, D. J. Edwards, and B. Shen. 2000. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC 15003 supporting functional interactions between nonribosomal peptide synthetases and polyketide synthase. Chem. Biol. 7:623-642. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Moreno, M. A., C. Vallin, and F. Malpartida. 1997. Streptothricin biosynthesis is catalyzed by enzymes related to nonribosomal peptide bond formation. J. Bacteriol. 179:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frey, P. A. 1993. Lysine 2,3-aminomutase: is adenosylmethionine a poor man's adenosylcobalamin? FASEB J. 7:662-670. [DOI] [PubMed] [Google Scholar]
- 20.Gould, S. J., and K. J. Martinkus. 1981. Biosynthesis of streptothricin F. 1. Observing the interaction of primary and secondary metabolism with [1,2-13C2]acetate. J. Am. Chem. Soc. 103:2871-2872. [Google Scholar]
- 21.Gould, S. J., and K. J. Martinkus. 1981. Studies of nitrogen metabolism using carbon-13 NMR spectroscopy. 2. Incorporation of l-[guanido-13C, 15N2]arginine and dl-[guanido-13C, 2-15N]arginine into streptothricin F. J. Am. Chem. Soc. 103:4639-4640. [Google Scholar]
- 22.Gould, S. J., and D. A. Minott. 1992. Biosynthesis of capreomycin. 1. Incorporation of arginine. J. Org. Chem. 57:5214-5217. [Google Scholar]
- 23.Grammel, N., K. Pankevych, J. Demydchuk, K. Lambrecht, H. P. Saluz, and H. Krugel. 2002. A beta-lysine adenylating enzyme and a beta-lysine binding protein involved in poly beta-lysine chain assembly in nourseothricin synthesis in Streptomyces noursei. Eur. J. Biochem. 269:347-357. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, M. D., S. J. Gould, and T. M. Zabriskie. 2002. Studies on the formation and incorporation of streptolidine in the biosynthesis of the peptidyl nucleoside antibiotic streptothricin F. J. Org. Chem. 67:2934-2941. [DOI] [PubMed] [Google Scholar]
- 25.Jenne, A., J. S. Hartig, N. Piganeau, A. Tauer, D. A. Samarsky, M. R. Green, J. Davies, and M. Famulok. 2001. Rapid identification and characterization of hammerhead-ribozyme inhibitors using fluorescence-based technology. Nat. Biotechnol. 19:56-61. [DOI] [PubMed] [Google Scholar]
- 26.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513-15521. [DOI] [PubMed] [Google Scholar]
- 27.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics, p. 249-250. The John Innes Foundation, Norwich, England.
- 28.Konz, D., and M. A. Marahiel. 1999. How do peptide synthetases generate structural diversity? Chem. Biol. 6:R39-R48. [DOI] [PubMed] [Google Scholar]
- 29.Kramnik, I., W. F. Dietrich, P. Demant, and B. R. Bloom. 2000. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 97:8560-8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawn, S. D., S. T. Butera, and T. M. Shinnick. 2002. Tuberculosis unleashed: the impact of human immunodeficiency virus infection on the host granulomatous response to Mycobacterium tuberculosis. Microbes Infect. 4:635-646. [DOI] [PubMed] [Google Scholar]
- 31.Legrain, C., and V. Stalon. 1976. Ornithine carbamoyltransferase from Escherichia coli W. Purification, structure and steady-state kinetic analysis. Eur. J. Biochem. 63:289-301. [DOI] [PubMed] [Google Scholar]
- 32.Linde, R. G., II, N. C. Birsner, R. Y. Chandrasekaran, J. Clancy, R. J. Howe, J. P. Lyssikatos, C. P. MacLelland, T. V. Magee, J. W. Petitpas, J. P. Rainville, W.-G. Su, C. B. Vu, and D. A. Whipple. 1997. Cyclic homopentapeptides. 3. Synthetic modifications to the capreomycins and tuberactinomycins: compounds with activity against methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci. Bioorg. Med. Chem. Lett. 7:1149-1152. [Google Scholar]
- 33.Lyssikatos, J. P., S.-P. Chang, J. Clancy, J. P. Dirlam, S. M. Finegan, A. E. Girard, S. F. Hayashi, D. P. Larson, A. S. Lee, R. G. Linde II, C. P. MacLelland, J. W. Petitpas, S. B. Seibel, and C. B. Vu. 1997. Cyclic homopentapeptides. 2. Synthetic modifications of viomycin. Bioorg. Med. Chem. Lett. 7:1145-1148. [Google Scholar]
- 34.MacNeil, D. J., K. M. Bewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-69. [DOI] [PubMed] [Google Scholar]
- 35.Mansouri, K., and W. Piepersberg. 1991. Genetics of streptomycin production in Streptomyces griseus: nucleotide sequence of five genes, strFGHIK, including a phosphatase gene. Mol. Gen. Genet. 228:459-469. [DOI] [PubMed] [Google Scholar]
- 36.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 37.Martin, M. F., and P. Liras. 1989. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 43:173-206. [DOI] [PubMed] [Google Scholar]
- 38.Martinkus, K. J., C. H. Tann, and S. J. Gould. 1983. The biosynthesis of streptothricin F. Part 4. The biosynthesis of the streptolidine moiety in streptothricin F. Tetrahedron 39:3493-3505. [Google Scholar]
- 39.Mason, J. R., and R. Cammack. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277-305. [DOI] [PubMed] [Google Scholar]
- 40.Murray, C. J. L., and J. A. Salomon. 1998. Modeling the impact of global tuberculosis control strategies. Proc. Natl. Acad. Sci. USA 95:13881-13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muth, W. L., and R. N. Costilow. 1974. Ornithine cyclase (deaminating). II. Properties of the homogeneous enzyme. J. Biol. Chem. 249:7457-7462. [PubMed] [Google Scholar]
- 42.Muth, W. L., and R. N. Costilow. 1974. Ornithine cyclase (deaminating). III. Mechanism of the conversion of ornithine to proline. J. Biol. Chem. 249:7463-7467. [PubMed] [Google Scholar]
- 43.Pootoolal, J., M. G. Thomas, C. G. Marshall, J. M. Neu, B. K. Hubbard, C. T. Walsh, and G. D. Wright. 2002. Assembling the glycopeptide antibiotic scaffold: the biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. USA 99:8962-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quadri, L. E., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 45.Rogers, J., A. H. Chang, U. von Ahsen, R. Schroeder, and J. Davies. 1996. Inhibition of the self-cleavage reaction of the human hepatitis delta virus ribozyme by antibiotics. J. Mol. Biol. 259:916-925. [DOI] [PubMed] [Google Scholar]
- 46.Salowe, S. P., E. N. Marsh, and C. A. Townsend. 1990. Purification and characterization of clavaminate synthase from Streptomyces clavuligerus: an unusual oxidative enzyme in natural product biosynthesis. Biochemistry 29:6499-6508. [DOI] [PubMed] [Google Scholar]
- 47.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 48.Tam, A. H.-K., and D. C. Jordan. 1972. Laboratory production and 14C-labelling of viomycin. J. Antibiot. 25:524-529. [DOI] [PubMed] [Google Scholar]
- 49.Thomas, M. G., M. D. Burkart, and C. T. Walsh. 2002. Conversion of l-proline to pyrrolyl-2-carboxyl-S-PCP during undecylprodigiosin and pyoluteorin biosynthesis. Chem. Biol. 9:171-184. [DOI] [PubMed] [Google Scholar]
- 50.Thorpe, C., and J. J. Kim. 1995. Structure and mechanism of action of the acyl-CoA dehydrogenases. FASEB J. 9:718-725. [DOI] [PubMed] [Google Scholar]
- 51.Tiffany, K. A., D. L. Roberts, M. Wang, R. Paschke, A. W. Mohsen, J. Vockley, and J. J. Kim. 1997. Structure of human isovaleryl-CoA dehydrogenase at 2.6 Å resolution: structural basis for substrate specificity. Biochemistry 36:8455-8464. [DOI] [PubMed] [Google Scholar]
- 52.Townsend, C. A. 2002. New reactions in clavulanic acid biosynthesis. Curr. Opin. Chem. Biol. 6:583-589. [DOI] [PubMed] [Google Scholar]
- 53.van Wageningen, A. M. A., P. N. Kirkpatrick, D. H. Williams, B. Harris, R., J. K. Kershaw, N. J. Lennard, M. Jones, S. J. M. Jones, and P. Solenberg. 1998. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 5:155-162. [DOI] [PubMed] [Google Scholar]
- 54.Wang, M., and S. J. Gould. 1993. Biosynthesis of capreomycin. 2. Incorporation of l-serine, l-alanine, and l-2, 3-diaminopropionic acid. J. Org. Chem. 58:5176-5180. [Google Scholar]
- 55.Wank, H., J. Rogers, J. Davies, and R. Schroeder. 1994. Peptide antibiotics of the tuberactinomycin family as inhibitors of group I intron RNA splicing. J. Mol. Biol. 236:1001-1010. [DOI] [PubMed] [Google Scholar]
- 56.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.World Health Organization. 2002. World Health Organization's model list of essential medicines. W. H. O. Drug Information 16:139-151. [Google Scholar]