Abstract
Sixty-three recent Italian clinical isolates of Streptococcus pyogenes resistant to both erythromycin (MICs ≥ 1 μg/ml) and tetracycline (MICs ≥ 8 μg/ml) were genotyped for macrolide and tetracycline resistance genes. We found 19 isolates carrying the mef(A) and the tet(O) genes; 25 isolates carrying the erm(A) and tet(O) genes; and 2 isolates carrying the erm(A), tet(M), and tet(O) genes. The resistance of all erm(A)-containing isolates was inducible, but the isolates could be divided into two groups on the basis of erythromycin MICs of either >128 or 1 to 4 μg/ml. The remaining 17 isolates included 15 isolates carrying the erm(B) gene and 2 isolates carrying both the erm(B) and the mef(A) genes, with all 17 carrying the tet(M) gene. Of these, 12 carried Tn916-Tn1545-like conjugative transposons. Conjugal transfer experiments demonstrated that the tet(O) gene moved with and without the erm(A) gene and with the mef(A) gene. These studies, together with the results of pulsed-field gel electrophoresis experiments and hybridization assays with DNA probes specific for the tet(O), erm(A), and mef(A) genes, suggested a linkage of tet(O) with either erm(A) or mef(A) in erythromycin- and tetracycline-resistant S. pyogenes isolates. By amplification and sequencing experiments, we detected the tet(O) gene ca. 5.5 kb upstream from the mef(A) gene. This is the first report demonstrating the presence of the tet(O) gene in S. pyogenes and showing that it may be linked with another gene and can be moved by conjugation from one chromosome to another.
Since the mid-1990s Italy has seen an increase in erythromycin-resistant Streptococcus pyogenes isolates (MICs ≥ 1 μg/ml) (30). In past studies, a correlation between the pulsed-field gel electrophoresis (PFGE) pattern, macrolide phenotypes (inducible versus constitutive), and tetracycline resistance was observed (8, 22); and it was concluded that the spread of erythromycin-resistant S. pyogenes in Italy is not due to the spread of a single clone (22). In the study described in this report, 63 recent Italian clinical strains of S. pyogenes resistant to both erythromycin and tetracycline were genotyped for the presence of erm(A) [subclass erm(TR) (27); hereafter designated erm(A) according to the present nomenclature (24)] and erm(B), which encode rRNA methylases; mef(A), which encodes a macrolide efflux protein (2); tet(M) and tet(O), both of which encode tetracycline resistance ribosomal protection proteins; and tet(K) and tet(L), both of which encode efflux proteins. The erythromycin resistance genes, erm(A), erm(B), and mef(A), are well known to occur in S. pyogenes (15). Both tetracycline resistance genes tet(M) and tet(O) have previously been reported in Streptococcus pneumoniae, but tet(O) has been reported from only two geographic areas (17, 31). While the tet(M) gene has been found in S. pyogenes (1), in which it was first detected in the prototype composite element Tn3701 (14), which contains a Tn916-like transposon (3), the tet(O) gene has not previously been identified in this species.
MATERIALS AND METHODS
Bacterial strains.
Sixty-three distinct clinical strains of S. pyogenes, collected from several Italian laboratories between 1997 and 2000 and isolated from throat swab cultures of symptomatic patients, were used. Strain identification was confirmed by using bacitracin disks (Difco Laboratories, Becton Dickinson, Sparks, Md.) and a latex agglutination assay (Streptex; Wellcome Diagnostics, Dartford, United Kingdom). Two inclusion criteria were used: all isolates were resistant to both erythromycin (MIC ≥ 1 μg/ml) and tetracycline (MIC ≥ 8 μg/ml), and all isolates were different strains on the basis of their phenotypic and genotypic characteristics.
Antibiotics and susceptibility tests.
Erythromycin, tetracycline, minocycline, and clindamycin were purchased from Sigma Chemical Co., St. Louis, Mo. Clarithromycin was obtained from Abbott Laboratories (Abbott Park, Ill.), azithromycin was obtained from Pfizer Inc. (New York, N.Y.), and josamycin was obtained from ICN Biomedicals (Costa Mesa, Calif.). MICs were determined by the broth microdilution method according to the protocols of the National Committee for Clinical Laboratory Standards (19). S. pneumoniae ATCC 49619 was used for quality control. Kanamycin and chloramphenicol susceptibilities were determined by a standard NCCLS agar diffusion test (20) with commercial disks (Oxoid Ltd., Basingstoke, United Kingdom) containing 30 μg of either antibiotic. The following zone diameter breakpoints were used for kanamycin: susceptible, ≥18 mm; intermediate, 14 to 17 mm; and resistant, ≤13 mm. The following zone diameter breakpoints were used for chloramphenicol: susceptible, ≥21 mm; intermediate, 18 to 20 mm; and resistant, ≤17 mm.
Determination of macrolide resistance phenotype.
Test strains were assigned to the constitutive, the inducible, or the efflux-mediated macrolide resistance phenotypes on the basis of their patterns of susceptibility to macrolide-lincosamide-streptogramin B antibiotics and the triple-disk (erythromycin, clindamycin, and josamycin) test, as described previously (8).
PFGE and random amplified polymorphic DNA analysis.
SmaI PFGE patterns were determined and analyzed as described recently (22). Random amplified polymorphic DNA analysis was performed by established methods (9) with primers M13 and H2 (26) with 21 of the isolates to distinguish strains that fell into single groups on the basis of their phenotypic or genotypic characteristics and PFGE analysis.
Gene detection and amplification experiments.
PCRs with specific primer pairs (Table 1) were used to detect erythromycin resistance genes erm(A), erm(B), and mef(A); tetracycline resistance genes tet(K), tet(L), tet(M), and tet(O); and the integrase gene int-Tn, associated with the Tn916-Tn1545 family of conjugative transposons. DNA preparation and amplification and electrophoresis of PCR products were carried out by established procedures (10) and according to the conditions indicated for the use of the individual primer pairs. PCR assays for determination of the nature of the tet(O)-erm(A) linkage were performed with one primer specific for a region within the tet(O) gene and a second primer specific for a region within the erm(A) gene (Table 1). Other PCR assays for determination of the nature of the tet(O)-mef(A) linkage were performed with one primer specific for a region within the tet(O) gene and a second primer specific for a region within the mef(A) gene or upstream of the mef(A) gene in orf3 (25) (Table 1). The Ex Taq system (TaKaRa Bio, Shiga, Japan) was used in these amplification experiments.
TABLE 1.
Sequences of primers and a tet(O)-specific probe used
| Gene | Primer or probe designation | Sequence (5′-3′) | Product size (bp) | Reference |
|---|---|---|---|---|
| erm(A) | III8a | GCATGACATAAACCTTCA | 208 | 27 |
| III10 | AGGTTATAATGAAACAGA | |||
| erm(A) | TR1b | ATAGAAATTGGGTCAGGAAAAGG | 530 | 12 |
| TR2 | TTGATTTTTAGTAAAAAG | |||
| erm(B) | ERMB1 | GAAAAGGTACTCAACCAAATA | 639 | 28 |
| ERMB2 | AGTAACGGTACTTAAATTGTTTAC | |||
| mef(A) | MEFA1c | AGTATCATTAATCACTAGTGC | 348 | 28 |
| MEFA2 | TTCTTCTGGTACTAAAAGTGG | |||
| tet(K) | tetK-up | TATTTTGGCTTTGTATTCTTTCAT | 1,159 | 29 |
| tetK-rev | GCTATACCTGTTCCCTCTGATAA | |||
| tet(L) | tetL-up | ATAAATTGTTTCGGGTCGGTAAT | 1,077 | 29 |
| tetL-rev | AACCAGCCAACTAATGACAATGAT | |||
| tet(M) | TETM2 | GAACTCGAACAAGAGGAAAGC | 740 | 21 |
| TETM3 | ATGGAAGCCCAGAAAGGAT | |||
| tet(O) | TETO1d | AACTTAGGCATTCTGGCTCAC | 519 | 21 |
| TETO2 | TCCCACTGTTCCATATCGTCA | |||
| int-Tn | int forward | GCGTGATTGTATCTCACT | 1,046 | 5 |
| int reverse | GACGCTCCTGTTGCTTCT | |||
| orf3e | ORF3rev | GACCTACCTGAACAATACC | 16 | |
| orf4e | ORF4 | AATATGGGCAGGGCAAGCA | 4 | |
| orf4e | OMI8 | TGCTTGCCCTGCCCATATT | 4 | |
| orf7e | ORF7 | GAATCTTTGGTCAGACTTGG | 16 | |
| orf8e | ORF8 | CCCTCCAATCCACCAGCG | 16 | |
| tet(O) | TO1f | CAACGATTGCAGTAAAGAAATCTG | 17 |
The III8-III10 primer pair was used to detect the erm(A) gene.
The TR1-TR2 primer pair was used to obtain an erm(A)-specific probe.
The MEFA1-MEFA2 primer pair was used to detect the mef(A) gene and obtain a mef(A)-specific probe.
The TETO1-TETO2 primer pair was used to detect the tet(O) gene and obtain a tet(O)-specific probe.
Gene of transposon Tn1207.1, containing eight open reading frames and carrying mef(A) as orf4 (25).
tet(O)-specific probe.
Mating experiments.
S. pyogenes strains iB21, iB27, iC38, and iC41 were used as donors for the erm(A) and the tet(O) genes; strains m46 and m49 were used as donors for the mef(A) and the tet(O) genes; and strains c7 and c13 were used as donors for the erm(B) and the tet(M) genes. S. pyogenes 12RF, a clinical strain selected in the laboratory for rifampin (25 μg/ml) and fusidic acid (25 μg/ml) resistance and susceptible to both erythromycin (≤0.015 μg/ml) and tetracycline (≤0.015 μg/ml), and Enterococcus faecalis JH2-2, a laboratory strain resistant to rifampin and fusidic acid, were both used as recipients of the genes from the S. pyogenes donors. Conjugal transfer was performed on a membrane filter (32) or directly on the agar surface as described previously (17). The frequency of transfer was expressed as the number of transconjugants per recipient. JH2-2 transconjugants carrying only the tet(O) gene from the S. pyogenes donors were used as donors, and E. faecalis OG1-10 (pPD1), which is resistant to 1,000 μg of streptomycin per ml (23), was used as a recipient in additional matings. Similarly, the S. pyogenes transconjugants were used as donors to the recipient S. pyogenes strain 12RF, which had been selected for resistance to streptomycin (500 μg/ml) and nalidixic acid (10 μg/ml). Selected transconjugants and donors were verified as carrying the tet(O) gene by the PCR assay (Table 1). Mating experiments were done a minimum of three times.
Plasmid isolation.
Plasmid isolation was performed as described previously (23).
Southern blotting and hybridization.
Total DNA was electrophoresed through a 1% agarose gel, transferred to a Zeta-Probe nylon membrane (Bio-Rad Laboratories, Richmond, Calif.) by capillary transfer, and hybridized with an [α-32P]dCTP-labeled DNA probe specific for the tet(O) gene (Table 1). DNA fragments generated by PFGE analysis were transferred to nylon membranes and hybridized with [α-32P]dCTP-labeled probes specific for the erm(A), mef(A), and tet(O) genes. These probes were obtained by PCR with the oligonucleotide primers reported in Table 1.
DNA sequence analysis.
Amplicon sequencing was performed bidirectionally by using the ABC Prism sequencer (Perkin-Elmer Italia, Monza, Italy) with dye-labeled terminators. Sequences were analyzed by using the Sequence Navigator software package (Perkin-Elmer).
RESULTS
Phenotypic and genotypic characterization of the strains.
The phenotypic and genotypic characteristics of the 63 S. pyogenes are listed in Table 2. The isolates carrying the erm(A) gene were inducibly resistant and could be divided into two groups on the basis of erythromycin MICs of >128 or 1 to 4 μg/ml. Twenty-five of the 27 erm(A)-positive isolates carried the tet(O) gene, and the remaining 2 isolates carried both the tet(O) and the tet(M) genes. Erythromycin MICs were 2 to 16 μg/ml for the 19 isolates carrying the mef(A) gene as the only erythromycin resistance determinant; they remained susceptible to clindamycin and josamycin also after induction, and all carried the tet(O) gene. The erythromycin MICs for the isolates carrying the erm(B) gene or both the erm(B) gene and the mef(A) gene were high (>128 μg/ml), and all carried the tet(M) gene. Of the 17 erm(B)-positive isolates, all were chloramphenicol susceptible; 12 isolates were kanamycin resistant and carried the int-Tn gene, indicating the presence of a conjugative transposon related to the Tn916-Tn1545 family. None of the 63 isolates carried either the tet(K) or the tet(L) gene, and plasmids were not observed.
TABLE 2.
Associations of erythromycin resistance genes erm(A), erm(B), and mef(A) and tetracycline resistance genes tet(M) and tet(O) in erythromycin- and tetracycline-resistant S. pyogenes strains, also characterized for their susceptibilities to clindamycin, clarithromycin, azithromycin, josamycin, and minocycline
| No. of strains tested | Macrolide resistance genotype | No. of strains with the tetracycline resistance gene:
|
MIC range (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tet(M) | tet(O) | tet(M) + tet(O) | ERY | TET | CLI | CLI (ind.)b | CLR | AZM | JOS | JOS (ind.)b | MIN | ||
| 15 | erm(B) | 15 | >128 | 32-64 | >128 | >128 | >128 | >128 | >128 | >128 | 4-16 | ||
| 2 | erm(B) + mef(A) | 2 | >128 | 32-64 | >128 | >128 | >128 | >128 | >128 | >128 | 8 | ||
| 17 | erm(A) | 15 | 2 | >128 | 32-64 | 0.03-0.25 | >128 | >128 | >128 | ≤0.03-0.12 | >128 | 2-8 | |
| 10 | erm(A) | 10 | 1-4 | 64->128 | ≤0.03-0.06 | >128 | 0.5-8 | 4-8 | ≤0.03-0.12 | 4-16 | 2-8 | ||
| 19 | mef(A) | 19 | 2-16 | 32-128 | ≤0.03-0.12 | ≤0.03-0.12 | 2-16 | 2-16 | ≤0.03-0.12 | ≤0.03-0.12 | 8-16 | ||
ERY, erythromycin; TET, tetracycline; CLI, clindamycin; CLR, clarithromycin; AZM, azithromycin; JOS, josamycin; MIN, minocycline.
ind., after induction by pregrowth in 0.05 μg of erythromycin per ml.
Conjugal transfer of erythromycin and tetracycline resistance genes.
Eight isolates with different combinations of macrolide and tetracycline resistance genes were used as donors in mating experiments. The results are summarized in Table 3. Seven of the eight isolates were able to transfer macrolide, tetracycline, or macrolide and tetracycline resistance to S. pyogenes and/or E. faecalis recipients at detectable frequencies under the conditions used. The frequencies of transfer from the donors with the erm(B) tet(M) and erm(A) tet(O) genotypes were lower when tetracycline was used for selection than when erythromycin was used for selection, regardless of which recipient was used; one exception was when donor iB27 and recipient JH2-2 were mated. The two isolates with the mef(A) tet(O) genotype did not transfer their genes to the E. faecalis recipient at measurable frequencies; however, they could transfer these genes to the S. pyogenes recipient at virtually the same frequency irrespective of whether tetracycline or erythromycin was used for selection, and all transconjugants carried both the mef(A) and the tet(O) genes. In contrast, strain iC41 transferred genes only to the E. faecalis recipient. With donors iB21 and iB27, segregation between the erm(A) and tet(O) genes was observed with both S. pyogenes and E. faecalis recipients. However, of 10 transconjugants obtained with donor iB21 and recipient 12RF, 9 had an erm(A) genotype and 1 had an erm(A) tet(O) genotype. Selected E. faecalis and S. pyogenes transconjugants carrying only the tet(O) gene were used as donors to examine whether they could transfer the tet(O) gene without the presence of the erm(A) gene. Both types of transconjugants could retransfer the tet(O) gene to the same species of recipient at the same frequencies shown in Table 3 (data not shown). Plasmids were not seen in the S. pyogenes transconjugants, and tet(O)-specific probe TO1 (Table 1) hybridized to the chromosomal DNA. No additional plasmids were seen in the E. faecalis OG1-10(pPD1) recipient, which carries a cryptic plasmid.
TABLE 3.
Conjugal transfer of erythromycin resistance genes erm(A) and mef(A) and/or tetracycline resistance gene tet(O) from erythromycin- and tetracycline-resistant S. pyogenes donors to susceptible S. pyogenes or E. faecalis recipients
| Donor | Recipient | Selection for resistance to:a | Transfer frequency | Transconjugants
|
|||
|---|---|---|---|---|---|---|---|
| Strain | Genotype | Genotype | MIC (μg/ml)a
|
||||
| ERY | TET | ||||||
| c7 | erm(B) tet(M) | 12RF | ERY | 1.3 × 10−7 | erm(B) tet(M) | >128 | 32 |
| c7 | erm(B) tet(M) | 12RF | TET | NDb | |||
| c7 | erm(B) tet(M) | JH2-2 | ERY | 7.1 × 10−8 | erm(B) tet(M) | >128 | 32 |
| c7 | erm(B) tet(M) | JH2-2 | TET | ND | |||
| c13 | erm(B) tet(M) | 12RF | ERY | 7.4 × 10−9 | erm(B) tet(M) | >128 | 32 |
| c13 | erm(B) tet(M) | 12RF | TET | 1.8 × 10−10 | erm(B) tet(M) | >128 | 32 |
| c13 | erm(B) tet(M) | JH2-2 | ERY | 1.9 × 10−7 | erm(B) | >128 | 0.25 |
| c13 | erm(B) tet(M) | JH2-2 | TET | ND | |||
| iB21c | erm(A) tet(O) | 12RF | ERY | 9 × 10−9 | erm(A) | >128 | ≤0.125 |
| 1 × 10−9 | erm(A) tet(O) | >128 | 64 | ||||
| iB21 | erm(A) tet(O) | 12RF | TET | ND | |||
| iB21 | erm(A) tet(O) | JH2-2 | ERY | 3.8 × 10−7 | erm(A) | >128 | 0.25 |
| iB21 | erm(A) tet(O) | JH2-2 | TET | 5 × 10−9 | tet(O) | ≤0.125 | 64 |
| iB27 | erm(A) tet(O) | 12RF | ERY | 2.5 × 10−6 | erm(A) | >128 | ≤0.125 |
| iB27 | erm(A) tet(O) | 12RF | TET | 7 × 10−7 | tet(O) | ≤0.125 | 64 |
| iB27 | erm(A) tet(O) | JH2-2 | ERY | 5.2 × 10−7 | erm(A) | >128 | 0.25 |
| iB27 | erm(A) tet(O) | JH2-2 | TET | 5 × 10−7 | tet(O) | ≤0.125 | 64 |
| iC38 | erm(A) tet(O) | 12RF | ERY | ND | |||
| iC38 | erm(A) tet(O) | 12RF | TET | ND | |||
| iC38 | erm(A) tet(O) | JH2-2 | ERY | ND | |||
| iC38 | erm(A) tet(O) | JH2-2 | TET | ND | |||
| iC41 | erm(A) tet(O) | 12RF | ERY | ND | |||
| iC41 | erm(A) tet(O) | 12RF | TET | ND | |||
| iC41 | erm(A) tet(O) | JH2-2 | ERY | 3.6 × 10−7 | erm(A) | >128 | 0.25 |
| iC41 | erm(A) tet(O) | JH2-2 | TET | ND | |||
| m46 | mef(A) tet(O) | 12RF | ERY | 6 × 10−4 | mef(A) tet(O) | 16 | 64 |
| m46 | mef(A) tet(O) | 12RF | TET | 5.8 × 10−4 | mef(A) tet(O) | 16 | 64 |
| m46 | mef(A) tet(O) | JH2-2 | ERY | ND | |||
| m46 | mef(A) tet(O) | JH2-2 | TET | ND | |||
| m49 | mef(A) tet(O) | 12RF | ERY | 1.3 × 10−7 | mef(A) tet(O) | 16 | 128 |
| m49 | mef(A) tet(O) | 12RF | TET | 2.1 × 10−7 | mef(A) tet(O) | 16 | 128 |
| m49 | mef(A) tet(O) | JH2-2 | ERY | ND | |||
| m49 | mef(A) tet(O) | JH2-2 | TET | ND | |||
ERY, erythromycin; TET, tetracycline. For the selection of transconjugants, erythromycin and tetracycline were used at concentrations of 1 and 10 μg/ml, respectively.
ND, no detectable transfer under conditions used.
Of 10 transconjugants obtained, 9 had the genotype erm(A) and one had the genotype erm(A) tet(O).
PFGE analysis and hybridization experiments.
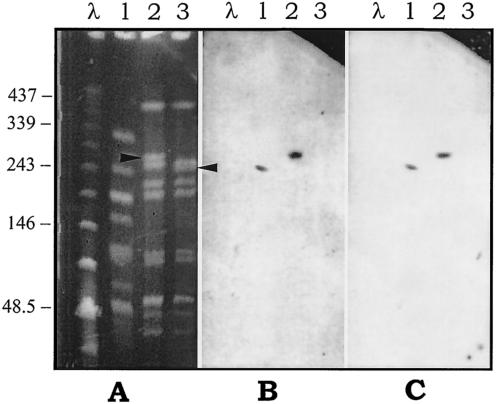
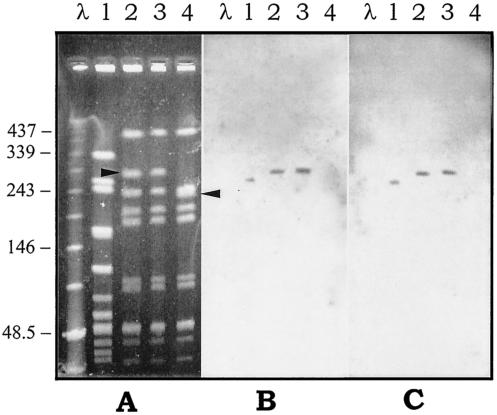
Donor iB21, recipient 12RF, and a 12RF transconjugant that was selected on erythromycin and that had an erm(A) tet(O) genotype were chosen for PFGE analysis. Compared with the PFGE profile of the recipient, the transconjugant exhibited a two-band difference resulting from the disappearance of a ca. 230-kb fragment and the appearance of a new one of ca. 260 kb (Fig. 1A). An erm(A)-specific probe (Fig. 1B) and a tet(O)-specific probe (Fig. 1C) both hybridized with an identical fragment from the donor and with an identical fragment (the new one) from the transconjugant. A similar experiment was done by mating donor m46 carrying the mef(A) and the tet(O) genes and recipient 12RF by using two 12RF transconjugants obtained by selecting one for erythromycin resistance and one for tetracycline resistance, but with both showing a mef(A) tet(O) genotype. The two transconjugants had identical PFGE patterns that differed from the PFGE pattern of the recipient by two bands resulting from the disappearance of a ca. 230-kb fragment (the same fragment described above) and the appearance of a new one of ca. 290 kb (Fig. 2A). A mef(A)-specific probe (Fig. 2B) and a tet(O)-specific probe (Fig. 2C) both hybridized with an identical fragment of the donor and with an identical fragment (the new one) of the transconjugants.
FIG. 1.
PFGE patterns of SmaI-digested genomic DNA of the S. pyogenes strains involved in the intraspecific mating iB21 × 12RF (A) and hybridization with the erm(A) specific probe (B) or the tet(O)-specific probe (C). Lanes 1, donor strain iB21; lanes 2, transconjugant selected for erythromycin resistance with an erm(A) tet(O) genotype; lanes 3, recipient strain 12RF. Bacteriophage lambda DNA concatemers (Bio-Rad) were used as molecular size markers (lanes λ). The arrowheads indicate a ca. 230-kb fragment of the recipient (lane 3) that disappeared and a ca. 260-kb new fragment that appeared in the transconjugant (lane 2). The numbers on the left are molecular sizes (in kilobases).
FIG. 2.
PFGE patterns of SmaI-digested genomic DNA of the S. pyogenes strains involved in the intraspecific mating m46 × 12RF (A) and hybridization with the mef(A)-specific probe (B) or the tet(O)-specific probe (C). Lanes 1, donor strain m46; lanes 2, transconjugant selected for erythromycin resistance with a mef(A) tet(O) genotype; lanes 3, transconjugant selected for tetracycline resistance with a mef(A) tet(O) genotype; lanes 4, recipient strain 12RF. Bacteriophage lambda DNA concatemers (Bio-Rad) were used as molecular size markers (lanes λ). The arrowheads indicate a ca. 230-kb fragment of the recipient (lane 4) that disappeared and a ca. 290-kb new fragment that appeared in the transconjugants (lanes 2 and 3). The numbers on the left are molecular sizes (in kilobases).
DNA amplification and sequencing.
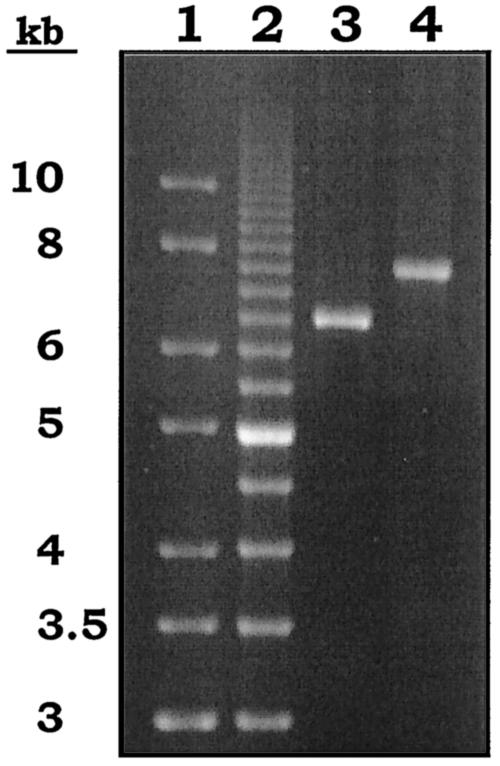
The PCR assay did not yield products with the primers specific for the tet(O) and the erm(A) genes. In contrast, PCR products were obtained by using the primers specific for the tet(O) gene and the mef(A) or the orf3 gene (Fig. 3). DNA sequencing confirmed the presence of the tet(O) gene at the left ends of both amplicons and of the orf3 and the mef(A) genes, respectively, at their right ends. On the basis of the results of this work, the tet(O) gene is ca. 5.5 kb upstream from mef(A).
FIG. 3.
Gel electrophoresis of long PCR products obtained from strain m46 [genotype mef(A) tet(O)] by using TETO1 as the tet(O)-specific primer (left end) and ORF3rev (lane 3; product size, ca. 6.5 kb) or OM18 (lane 4; product size, ca. 7.5 kb) to target orf3 or mef(A) at the right end. GeneRuler (M-Medical Genenco, Cornaredo, Italy) (lane 1) and a 500-bp molecular size ruler (Bio-Rad) (lane 2) were used as molecular size markers.
DISCUSSION
The increase in the prevalence of erythromycin-resistant S. pyogenes strains carrying the erm(A), erm(B), and/or mef(A) gene has been the subject of a number of recent reports (8, 11, 15, 27). In contrast, less work has been done to characterize the mechanism of tetracycline resistance in this species in which only tet(M) has been the commonly identified gene. Nevertheless, other species of streptococci which carry the tet(O) gene, which codes for another tetracycline resistance ribosomal protection protein, or the tet(K) and the tet(L) genes, which code for efflux-mediated tetracycline resistance (1), have been identified. Therefore, the potential exists for S. pyogenes to acquire other tetracycline resistance genes. In this study we found that 46 (73%) of 63 tetracycline- and erythromycin-resistant S. pyogenes isolates carried the tet(O) gene and the erm(A) or the mef(A) gene. In contrast, all the remaining isolates carried the tet(M) and the erm(B) genes, with 71% having conjugative transposons related to the Tn916-Tn1545 family on the basis of detection of the int-Tn gene.
A linkage between the erm(B) and the tet(M) genes has been well established in a variety of gram-positive cocci (1, 3), whereas a linkage involving tet(O) has not previously been reported. The differential transfer of the tet(O) and the erm(A) genes in the mating experiments, the ability to transfer genes to both S. pyogenes and E. faecalis, and the appearance of a single insertion in the transconjugant receiving both tet(O) and erm(A) suggest that these two genes are associated with conjugative elements. Moreover, Southern blotting analysis indicated that the single extra band hybridized with both a tet(O)-specific probe and an erm(A)-specific probe. The fact that the acquisition of erm(A) and tet(O) conferred to the transconjugant a PFGE pattern denoting the insertion of new DNA into an existing restriction fragment is consistent with a chromosomal location of the two genes. The erm(A) gene has previously been described on transposon Tn554 (24) and has more recently been shown to transfer from S. pyogenes into a variety of recipients (6), while the tet(O) gene has been found on plasmids or in the chromosome (1, 13, 31, 33). These strains are also interesting because of a recent report showing an association between the erm(A) and the erm(B) genes and the prtF1 gene, which encodes a protein required for streptococcal invasion of eukaryotic cells (6).
The association between the mef(A) gene and the tet(O) gene is suggested by the mating experiments and by PFGE and hybridization studies. Again, the PFGE patterns of the transconjugants with a mef(A) tet(O) genotype showed a single insertion of new DNA, consistent with an association and a chromosomal location of the two genes. Likewise, Southern blotting analysis indicated that the single extra band hybridized with both a tet(O)-specific probe and a mef(A)-specific probe. The mef(A) gene has been shown to be associated with both conjugative and nonconjugative elements as well as composite elements (15, 18, 25).
The fact that amplification and sequencing experiments ruled out a close proximity of tet(O) and erm(A) was not surprising, considering that the two genes were successfully cotransferred in a single mating experiment, with selection only for erythromycin resistance, and to only 1 of 10 transconjugants. In contrast, the finding of a linkage between tet(O) and mef(A), which is the first documented linkage between tet(O) and another gene, is consistent with the fact that the two genes were always cotransferred to the S. pyogenes recipient at the same frequency whether tetracycline or erythromycin was used for selection. The mef(A) gene has been shown to be carried by Tn1207.1, a chromosomal, defective, nonconjugative transposon of ca. 7.2 kb (25). In clinical isolates of S. pyogenes, Tn1207.1 has been reported to be part of a larger conjugative transposon, Tn1207.3 (M. Santagati, F. Iannelli, C. Messina, M. R. Oggioni, S. Stefani, and G. Pozzi, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-2014, 2001). The reported size of Tn1207.3 (ca. 52 kb) appears to be consistent with the disappearance of a ca. 230-kb fragment of the recipient and the appearance of a new fragment of ca. 290 kb in the transconjugants (Fig. 2).
The 63 Italian S. pyogenes isolates characterized in this study illustrate the ongoing evolution that is occurring as these bacteria cope with the ever-changing landscape in the clinical environment. This is the first study showing that the tet(O) gene can be mobile also when it is found in the chromosome. One can predict that if tet(O) is now associated with a conjugative element, then this gene is likely to spread to other streptococci and other gram-positive and gram-negative species, similar to what has been found with the host range of tet(M) (1).
Acknowledgments
The active contributions of Patrizia Bagnarelli, Aldo Manzin, Stefano Menzo, Kayode K. Ojo, and Nicole van Kirk in DNA sequence analysis and of Marina Moroni and Massimiliano Zampini in other experiments are gratefully acknowledged.
This work was supported in part by MIUR (Italian Ministry of Education, University and Research) grant MUVAR01302 and by NIH grant U24 AI50139-01A1.
REFERENCES
- 1.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 3.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 4.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. G. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptoccus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Facinelli, B., C. Spinaci, G. Magi, E. Giovanetti, and P. E. Varaldo. 2001. Association between erythromycin resistance and ability to enter human respiratory cells in group A streptococci. Lancet 358:30-33. [DOI] [PubMed] [Google Scholar]
- 7.Giovanetti, E., G. Magi, A. Brenciani, C. Spinaci, R. Lupidi, B. Facinelli, and P. E. Varaldo. 2002. Conjugative transfer of the erm(A) gene from erythromycin-resistant Streptococcus pyogenes to macrolide-susceptible S. pyogenes, Enterococcus faecalis, and Listeria innocua. J. Antimicrob. Chemother. 50:249-252. [DOI] [PubMed] [Google Scholar]
- 8.Giovanetti, E., M. P. Montanari, M. Mingoia, and P. E. Varaldo. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruteke, P., A. van Belkum, L. M. Schouls, W. D. H. Hendriks, F. A. G. Reubsaet, J. Dokter, H. Boxma, and H. A. Verbrugh. 1996. Outbreak of group A streptococci in a burn center: use of pheno- and genotypic procedures for strain tracking. J. Clin. Microbiol. 34:114-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hynes, W. L., J. J. Ferretti, M. S. Gilmore, and R. A. Segarra. 1992. PCR amplification of streptococcal DNA using crude cell lysates. FEMS Microbiol. Lett. 94:139-142. [DOI] [PubMed] [Google Scholar]
- 11.Kataja, J., P. Huovinen, the Macrolide Resistance Study Group, and H. Seppälä. 2000. Erythromycin resistance genes in group A streptococci of different geographical origins. J. Antimicrob. Chemother. 46:789-792. [DOI] [PubMed] [Google Scholar]
- 12.Kataja, J., H. Seppälä, M. Skurnik, H. Sarkkinen, and P. Huovinen. 1998. Different erythromycin resistance mechanisms in group C and group G streptococci. Antimicrob. Agents Chemother. 42:1493-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeBlanc, D. J., L. N. Lee, B. M. Titmas, C. J. Smith, and F. C. Tenover. 1988. Nucleotide sequence analysis of tetracycline resistance gene tetO from Streptococcus mutans DL5. J. Bacteriol. 170:3618-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Bouguenec, C., G. de Cespedes, and T. Horaud. 1990. Presence of chromosomal elements resembling the composite structure Tn3701 in streptococci. J. Bacteriol. 172:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 16.Luna, V. A., M. Heiken, K. Judge, C. Ulep, N. Van Kirk, H. Luis, M. Bernardo, J. Leitao, and M. C. Roberts. 2002. Distribution of mef(A) in gram-positive bacteria from healthy Portuguese children. Antimicrob. Agents Chemother. 46:2513-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luna, V. A., and M. C. Roberts. 1998. The presence of the tetO gene in a variety of tetracycline-resistant Streptococcus pneumoniae serotypes from Washington State. J. Antimicrob. Chemother. 42:613-619. [DOI] [PubMed] [Google Scholar]
- 18.Luna, V. A., P. Coates, E. A. Eady, J. Cove, T. T. H. Nguyen, and M. C. Roberts. 1999. A variety of gram-positive bacteria carry mobile mef genes. J. Antimicrob. Chemother. 44:19-25. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests, 7th ed. Approved standard M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.Olsvik, B., I. Olsen, and F. C. Tenover. 1995. Detection of tet(M) and tet(O) using the polymerase chain reaction in bacteria isolated from patients with periodontal disease. Oral Microbiol. Immunol. 10:87-92. [DOI] [PubMed] [Google Scholar]
- 22.Ripa, S., C. Zampaloni, L. A. Vitali, E. Giovanetti, M. P. Montanari, M. Prenna, and P. E. Varaldo. 2001. SmaI macrorestriction analysis of Italian isolates of erythromycin-resistant Streptococcus pyogenes and correlations with macrolide-resistance phenotypes. Microb. Drug Resist. 7:65-71. [DOI] [PubMed] [Google Scholar]
- 23.Roberts, M. C., and G. E. Kenny. 1987. Conjugal transfer of transposon Tn916 from Streptococcus faecalis to Mycoplasma hominis. J. Bacteriol. 169:3836-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppälä. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santagati, M., F. Iannelli, M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seppälä, H., Q. He, M. Österblad, and P. Huovinen. 1994. Typing of group A streptococci by random amplified polymorphic DNA analysis. J. Clin. Microbiol. 32:1945-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trzcinski, K., B. S. Cooper, W. Hryniewicz, and C. G. Dowson. 2000. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus. aureus. J. Antimicrob. Chemother. 45:763-770. [DOI] [PubMed] [Google Scholar]
- 30.Varaldo, P. E., E. A. Debbia, G. Nicoletti, D. Pavesio, S. Ripa, G. C. Schito, G. Tempera, and the Artemis-Italy Study Group. 1999. Nationwide survey in Italy of treatment of Streptococcus pyogenes pharyngitis in children: influence of macrolide resistance on clinical and microbiological outcomes. Clin. Infect. Dis. 29:869-873. [DOI] [PubMed] [Google Scholar]
- 31.Widdowson, C. A., K. P. Klugman, and D. Hanslo. 1996. Identification of the tetracycline resistance gene, tet(O), in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2891-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willetts, N. 1988. Conjugation. Methods Microbiol. 21:49-77. [Google Scholar]
- 33.Zilhao, R., B. Papadopoulou, and P. Courvalin. 1988. Occurrence of the Campylobacter resistance gene tetO in Enterococcus and Streptococcus spp. Antimicrob. Agents Chemother. 32:1793-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]