Abstract
Human β-defensin 3 (hBD3) is a highly basic 45-amino-acid protein that acts both as an antimicrobial agent and as a chemoattractant molecule. Although the nature of its antimicrobial activity is largely electrostatic, the importance of the molecular structure on this activity is poorly understood. Two isoforms of hBD3 were synthesized: the first with native disulfide linkages and the second with nonnative linkages. In a third synthetic peptide, all cysteine residues were replaced with α-aminobutyric acid, creating a completely linear peptide. A series of six small, linear peptides corresponding to regions of hBD3 with net charges ranging from +4 to +8 (at pH 7) and lengths ranging from 9 to 20 amino acids were also synthesized. The linear full-length peptide showed the highest microbicidal activity against Escherichia coli and Staphylococcus aureus, while all three full-length forms showed equal activity against Candida albicans. The linear peptide also showed high activity against Enterococcus faecium and Pseudomonas aeruginosa. Peptides corresponding to the C terminus showed higher activities when tested against E. coli, with the most active peptides being the most basic. However, only the peptide corresponding to the N terminus of hBD3 showed any activity against S. aureus and C. albicans. Further, N-terminal deletion mutants of native hBD3 showed diminished activities against S. aureus. Thus, the antimicrobial properties of hBD3 derivatives are determined by both charge and structure.
Defensins are small, cationic peptides, classified by their pattern of conserved cysteines, that are primarily known for their antimicrobial properties (6, 26). They also act as chemoattractant agents for monocytes and dendritic cells in mammals (36). Defensins in mammals are categorized into three families—α-, β-, and θ-defensins—based on sequence homology and disulfide pairing. In humans, there are currently six α-defensins (known as neutrophil peptides 1 to 6 [hNP1 to hNP6]) and four β-defensins (hBD1 to hBD4). θ-Defensins were discovered in the rhesus monkey; the human homolog has not yet been described (32).
Various defensins differ substantially in their antimicrobial specificities. For example, hBD2 has considerable activity against gram-negative bacteria and the fungus Candida albicans but little to no activity against gram-positive bacteria (12). The recently discovered hBD4 showed an MIC of ∼4 μg/ml for Staphylococcus carnosus and Pseudomonas aeruginosa, with MICs of >100 μg/ml for Escherichia coli, Saccharomyces cerevisiae, Staphylococcus aureus, Streptococcus pneumoniae, and Burkholderia cepacia in conventional dilution assays (9). On the other hand, hNP1, hNP5, hBD1, and hBD3 display broad antimicrobial activity against gram-negative and gram-positive bacteria, fungi, and adenovirus (1, 7, 11, 13, 25).
Human defensins are of significant interest recently due to their antimicrobial and immunomodulatory activities, coupled with their inherent immunological compatibility. The practical therapeutic use of human defensins is impaired, however, by their size, complexity of disulfide pairing, and inflammatory actions. Also, the antimicrobial activity of defensins is attenuated significantly by elevated ionic strength. Studies of peptide fragments and cyclic analogs of defensins have shown some promise for engineering novel, salt-insensitive antimicrobial agents (22, 38).
The defensin hBD3 was found to have considerable salt-independent potency toward gram-positive bacteria (8, 13, 19). Recently, it has been shown that the antimicrobial activity of hBD3 is independent of its pattern of disulfide pairings (35a). Furthermore, replacement of cysteine residues in hBD3 with α-aminobutyric acid rendered the peptide chemotactically inactive, yet with enhanced antimicrobial activity (35a). In an attempt to further characterize this effect, as well as to minimize the region necessary for antimicrobial activity of the linearized hBD3 peptide, we synthesized a set of peptides, derived from the hBD3 sequence, of various sizes and with different disulfide pairings and charges, and we tested them against E. coli, P. aeruginosa, Enterococcus faecium, S. aureus, and C. albicans.
MATERIALS AND METHODS
Peptide synthesis.
Peptides (all sequences shown in Table 1) hBD3 (full-length hBD3 with native disulfide pairings Cys11-Cys40, Cys18-Cys33, and Cys23-Cys41), hBD3A (full-length hBD3 with disulfide pairings Cys11-Cys41, Cys18-Cys40, and Cys23-Cys33), ABU-hBD3 (full-length hBD3 with cysteine residues replaced by α-aminobutyric acid), hBD3Δ8 (residues 8 to 45 of hBD3 with native disulfide pairings), and hBD3Δ10 (residues 10 to 45 of hBD3 with native disulfide pairings) were synthesized as described elsewhere (35a). In brief, the peptides were prepared in solid phase by using a custom-modified chemistry tailored based on the published in situ neutralization protocols for Boc solid-phase peptide synthesis (28). All peptides were purified to homogeneity by reversed phase high-pressure liquid chromatography (RP-HPLC), and their molecular weights verified by electrospray ionization mass spectrometry (ESI-MS). The peptides were refolded and oxidized via a rapid sixfold dilution of fully reduced peptides dissolved at 1.5 mg/ml in 6 M GuHCl into a final buffer solution containing 0.1 M NaHCO3, 1 M guanidine HCl, 3 mM cysteine, and 0.3 mM cystine (pH 8.1). The folding reaction typically proceeded at room temperature overnight in a sealed vial with gentle stirring.
TABLE 1.
Peptides used in this study
| Peptide | Sequencea | Disulfide connectivity | Net charge | Length (amino acids) |
|---|---|---|---|---|
| hBD3 | GIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK | 1-5, 2-4, 3-6 | +11 | 45 |
| hBD3A | GIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK | 1-6, 2-5, 3-4 | +11 | 45 |
| hBD3Δ8 | KYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK | 1-5, 2-4, 3-6 | +11 | 38 |
| hBD3Δ10 | YCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK | 1-5, 2-4, 3-6 | +10 | 36 |
| ABU-hBD3 | GIINTLQKYYαRVRGGRαAVLSαLPKEEQIGKαSTRGRKααRRKK | +11 | 45 | |
| STRC06 | KEEQIGKSSTRGRKSSRRKK | +7 | 20 | |
| CHRG01 | KSSTRGRKSSRRKK | +8 | 14 | |
| CHRG02 | RGRKSSRRKK | +7 | 10 | |
| CHRG04 | RGRKSSRRK | +6 | 9 | |
| CHRG06 | KYYSRVRGGRSAVLSSLPK | +5 | 19 | |
| CHRG07 | GIINTLQKYYSRVRGGR | +4 | 17 |
α, α-aminobutyric acid.
In order to define disulfide bond linkages, the synthetic strategy using reversible protection of selected Cys residues by acetamidomethyl (Acm) groups was employed. For hBD3 with six Cys residues, there are a total of 15 possible isoforms with distinctive disulfide pairings. Protection of two of six Cys residues by Acm reduces the number of oxidation products from 15 to 3, thus significantly simplifying the separation and subsequent determination of the disulfide connectivity. Residues Cys11 and Cys41 were protected for peptides hBD3, hBD3Δ8, and hBD3Δ10, whereas residues Cys11 and Cys40 were protected for peptide hBD3A. Thus, six oxidation products, each containing two disulfide bridges, were obtained from two fully reduced Cys11(Acm)/Cys40(Acm) and Cys11(Acm)/Cys41(Acm) peptides.
Prior to carrying out the full-scale protocol, we subjected the fractions purified by preparative HPLC to enzymatic digestion with trypsin and chymotrypsin to determine the disulfide connectivity. Because mass spectrometric identification of tryptic fragments yielded a chemically identical, two-disulfide-bridged tryptic fragment of 1,846.9 Da, the ambiguity was resolved by subsequent enzymatic digestion with chymotrypsin. All of the common and unique fragments generated by trypsin and chymotrypsin were identified and accounted for by analytical RP-HPLC and ESI-MS. Deprotection and spontaneous oxidation of Cys11/Cys40 or Cys11/Cys41 was achieved through treatment with I2 at acidic pH according to the published protocols (31). Final products were verified by ESI-MS, yielding an observed mass value of 5,155.5 Da, which is in agreement with the expected value of 5,155.2 Da, calculated based on the average isotope compositions of folded hBD3. To demonstrate chromatographic purity, samples were analyzed on RP-HPLC.
Peptides STRC01, CHRG01, CHRG02, CHRG04, CHRG06, and CHRG07 were synthesized by the ICBR Protein Chemistry and Biomarkers Core Facility, University of Florida College of Medicine, Gainesville, Fla. All of the cysteine residues in the short peptides were replaced with serine to eliminate disulfide formation. The identity and purity of all peptides was confirmed by ESI-MS and RP-HPLC.
Antimicrobial assays.
The microorganisms assayed included E. coli ATCC 25922, S. aureus ATCC 29213, C. albicans 99788 (amphotericin B resistant), E. faecium 1438, and P. aeruginosa PAO1. Assays for antimicrobial activity are essentially those previously described for defensins (14). The microorganisms were grown to mid-logarithmic phase in tryptic soy broth and then diluted to 106 CFU/ml in 10 mM potassium phosphate-1% tryptic soy broth (pH 7.4). For salt dependence assays, either 150 mM NaCl or 1 mM MgCl2 plus 2 mM CaCl2 were included in the dilution buffer. Then, 100-μl portions of cells were incubated in the presence of different concentrations of peptides for 3 h at 37°C. The cells were then diluted serially in the same buffer, plated on Luria broth plates, and incubated for 18 h at 33°C, and the colonies were counted. Microbicidal activity was expressed as the ratio of colonies counted to the number of colonies on a control plate. The 90% lethal concentration (LC90) is the concentration of peptide at which 90% of the viable cells are killed. The results are shown in Table 2. Plates of dilutions with >500 colonies were not counted. All assays were done in triplicate, and error bars are indicated in Fig. 1 to 4. Generally, the differences in the number of colonies counted on identical plates (with between 20 and 500 colonies on each plate) varied between 20 and 30%.
TABLE 2.
LC90s of peptides
| Peptide | Meana LC90 (μg/ml) ± SD against:
|
||||
|---|---|---|---|---|---|
| E. coli | S. aureus | C. albicans | P. aeruginosa | E. faecium | |
| hBD3 | 6 ± 1.4 | 10 ± 4.5 | 17 ± 3.2 | ||
| hBD3A | 5 ± 0.8 | 14 ± 8.9 | 15 ± 5.1 | ||
| hBD3Δ8 | 6 ± 0.4 | 16 ± 5.4 | NDb | ||
| hBD3Δ10 | 6 ± 1.9 | 17 ± 10.2 | ND | ||
| ABU-hBD3 | 1 ± 0.2 | 5 ± 0.7 | 17 ± 3.1 | 4 ± 1.8 | 2 ± 0.5 |
| STRC06 | 20 ± 4.6 | >20 | >20 | ||
| CHRG01 | 1 ± 0.1 | >20 | >20 | 4 ± 0.4 | >20 |
| CHRG02 | 4 ± 0.3 | >20 | >20 | ||
| CHRG04 | 10 ± 2.8 | >20 | >20 | ||
| CHRG06 | 9 ± 1.8 | >20 | >20 | ||
| CHRG07 | 19 ± 3.4 | 17 ± 3.4 | 15 ± 2.1 | 2 ± 0.7 | 3 ± 1.6 |
Based on three independent assays.
ND, not done.
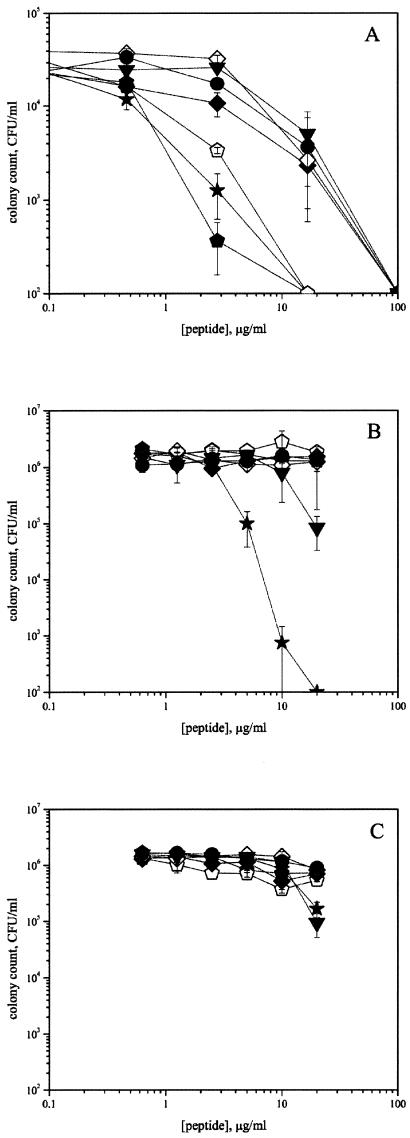
FIG. 1.
Antimicrobial activity of full-length hBD3 peptides. Symbols: ▪, hBD3; □, hBD3A; ★, ABU-hBD3. (A) E. coli; (B) S. aureus; (C) C. albicans.
FIG. 4.
Antimicrobial activity of selected peptides. Symbols: ★, ABU-hBD3; solid pentagons, CHRG01; ▾, CHRG07. (A) P. aeruginosa; (B) E. faecium.
RESULTS
Native hBD3 showed approximately the same antimicrobial activity against E. coli, S. aureus, and C. albicans as hBD3A (LC90s of ∼5 μg/ml for E. coli, ∼12 μg/ml for S. aureus, and ∼15 μg/ml for C. albicans) (Fig. 1). ABU-hBD3 somewhat showed higher activity against both E. coli and S. aureus (LC90s of ∼1 and ∼5 μg/ml, respectively), whereas it had about the same activity for C. albicans as native hBD3 (LC90 value of ∼17 μg/ml).
When tested against E. coli, peptides corresponding to the C-terminal region showed the highest antimicrobial activity (Fig. 2). Peptide CHRG01, with a net charge of +8, was as potent as ABU-hBD3 (LC90 of ∼1 μg/ml), and CHRG02 (net charge of +7) was almost as potent as CHRG01 and ABU-hBD3 (LC90 of ∼4 μg/ml). The other peptides, with net charges ranging from +4 to +7, showed decreased activity. Consequently, peptide CHRG07, with a net charge of +4, showed the least activity, reflected by an LC90 of ∼19 μg/ml. An exception to the trend seen (higher positive charge and C-terminal region) was found for STRC06 (net charge of +7), which was characterized by an LC90 of ∼20 μg/ml, an LC90 similar to that for CHRG07.
FIG. 2.
Antimicrobial activity of hBD3-derived peptides. Symbols: ★, ABU-hBD3; solid pentagons, CHRG01; open pentagons, CHRG02; ♦, CHRG04; ⋄, CHRG06; ▾, CHRG07; •, STRC06. (A) E. coli; (B) S. aureus; (C) C. albicans.
The only peptide that showed any antimicrobial activity against both pathogens in the concentration range tested was CHRG07 (net charge of +4). This peptide has considerable hydrophobic character and, in the native hBD3, forms an α-helix, as seen in the nuclear magnetic resonance solution structure (27). The hydrophobicity may play a role, since another peptide—CHRG06, with the same number of hydrophobic residues as CHRG07 (including the tyrosine residues)—shows activity against S. aureus only at concentrations of >20 μg/ml (data not shown).
The peptide CHRG07 contains seven additional residues compared to those found in CHRG06. To test the significance of these additional residues for antimicrobial activity, two truncated forms of hBD3 were synthesized, hBD3Δ8 (residues 8 to 45) and hBD3Δ10 (residues 10 to 45). Both peptides were oxidized selectively to form native disulfide pairings. As shown in Fig. 3, the truncation has no effect on activity against E. coli, while having an effect against S. aureus only at the highest concentration tested. Thus, whereas the N-terminal region likely plays a dominant role in mediating activity against S. aureus, there are probably multiple regions important for activity.
FIG. 3.
Antimicrobial activity of truncated hBD3. Symbols: ▪, hBD3; ▴, hBD3Δ8; ▵, hBD3Δ10; ★, ABU-hBD3; ▾, CHRG07. (A) E. coli; (B) S. aureus.
In order to investigate more closely the importance of electrostatic interactions for activity, we incubated peptides with E. coli and S. aureus in the presence of either 150 mM NaCl or the mixture of 1 mM MgCl2 and 2 mM CaCl2 (Table 3). The antimicrobial activity of ABU-hBD3 was resistant to both 150 mM NaCl and low concentrations of Mg and Ca for E. coli, but NaCl attenuated its activity against S. aureus. Under similar conditions, the activities of both hBD3 and hBD3A against E. coli were greatly diminished. The activities of both peptides, CHRG01 and CHRG07, in assays with either E. coli and S. aureus, were also decreased in the presence of NaCl or Mg and Ca.
TABLE 3.
Salt dependence of activity
| Peptide | Mean LC90 (μg/ml) ± SDa
|
|||
|---|---|---|---|---|
|
E. coli with:
|
S. aureus with:
|
|||
| 150 mM NaCl | 1 mM Mg and 2 mM Ca | 150 mM NaCl | 1 mM Mg and 2 mM Ca | |
| hBD3 | - | - | ND | ND |
| hBD3A | - | - | ND | ND |
| ABU-hBD3 | 11 ± 2.1 | 2 ± 0.5 | - | 6 ± 1.2 |
| CHRG01 | - | - | ND | ND |
| CHRG07 | - | - | - | - |
—, no measurable activity within the range of concentrations measured. ND, not done.
To extend the findings to additional pathogens, peptides CHRG01 and CHRG07, as well as ABU-hBD3, were tested against the gram-positive bacterium E. faecium and the gram-negative bacterium P. aeruginosa (Fig. 4). The activities of ABU-hBD3 against both bacteria were comparable to those observed earlier in assays with E. coli and S. aureus (LC90 of ca. 2 to 4 μg/ml). The peptide CHRG01 was active only against P. aeruginosa (LC90 of ∼4 μg/ml), with no activity seen against E. faecium, whereas the peptide CHRG07 was equally active against both bacterial species (LC90 of ca. 2 to 3 μg/ml).
DISCUSSION
The antimicrobial mechanism of defensins involves the disruption of negatively charged membranes, leakage of cellular contents, short-circuiting of the proton gradient and ultimately the destruction of the cell by osmolysis. Defensins have been shown to integrate and pass through such membranes, but the details of interactions with membranes vary between defensin molecules. Several mechanisms of antimicrobial activity of defensin have been postulated in the past. They involve distinct pore formation (5, 35) and local surface oligomerization (16, 21), as well as nonspecific electrostatic neutralization and voltage-dependent ion gating (4, 17, 23). Evidence indicates that intracellular components are also involved in these processes (10, 29).
The specificity of defensin interactions with microbial membranes is not well understood. While defensin molecules are attracted to negatively charged membranes typically found in microorganisms, additional factors may play roles in generating differential specificity. In particular, the structural features of these proteins likely contribute to their potency and specificity. Whereas all mammalian defensins share the same topology of a small β-sheet cross-linked by three disulfide bonds, only β-defensins contain an α-helix at the N terminus. Because of the small size of defensins, it is thought that the disulfides are critical in maintaining the three-dimensional fold and structure of the molecule. The current study has shown that, in the case of hBD3, disulfides are not completely necessary for antimicrobial activity against the microorganisms investigated. Further, under physiological salt conditions (150 mM NaCl), only ABU-hBD3 showed significant activity against E. coli (Table 3). Whereas it has been shown that ABU-hBD3 does not hold any single, stable tertiary structure in solution (35a), its regions may transiently accommodate the native fold without disulfide cross-linking. Also, the environment in a lipid membrane may allow alternate structures with similar functional activity.
HBD3 was discovered and isolated on the basis of its activity against S. aureus (13). However, it displays broad specificity against a number of microorganisms. Therefore, the hBD3 molecules can interact with a wide variety of membranes from multiple microbial species. It has been shown that the components of lipid membranes play a dominant role in discriminating interactions with different defensins. These components include lipopolysaccharides (3, 30), cardiolipin (18), teichoic acids (24), and sphingolipids (33). Also important in mitigating antimicrobial activity is ΔΨ (4, 20, 37). Perhaps the hBD3 molecule consists of separate regions capable of interacting with membrane components from multiple species.
We showed here that the activity of ABU-hBD3 can be mimicked by shorter peptides, although with altered specificities. Although the net charge of the peptide seems to play a role in its antibacterial activity against E. coli, this observation does not hold up with all microbial species. Other than the net charge, the features of hBD3 must therefore play roles in interacting with the membranes of S. aureus, P. aeruginosa, E. faecium, and C. albicans. This observation is clearly reflected for the peptide CHRG07. Amphilicity was cited as the key property of hNP-1 for its antimicrobial activity (15, 34). The presence of arginine residues and aromatic side chains was also shown to be important for antibacterial activity of small peptides (2, 27). The activity of CHRG07 might thus stem from the presence of two tyrosine residues and the hydrophobic N terminus, as observed for hBD3Δ8 and hBD3Δ10. The findings that the disulfides of hBD3 are irrelevant for antimicrobial activity under the conditions in the present study and that shorter peptides derived from the native sequence have full activity may help in designing a novel class of small-molecule antibiotics with broad specificity. Furthermore, the fact that hBD3 is a natural human protein shows promise for nonrejectable peptide-based antibiotics.
Acknowledgments
This research was sponsored in part by the Intramural AIDS Targeted Antiviral Program of the Office of the Director, National Institutes of Health (J.L.). This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-1240 (K.T.).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organization imply endorsement by the U.S. Government.
REFERENCES
- 1.Bensch, K. W., M. Raida, H. J. Magert, P. Schulz-Knappe, and W. G. Forssmann. 1995. hBD-1: a novel beta-defensin from human plasma. FEBS Lett. 368:331-335. [DOI] [PubMed] [Google Scholar]
- 2.Blondelle, S. E., and K. Lohner. 2000. Combinatorial libraries: a tool to design antimicrobial and antifungal peptide analogues having lytic specificities for structure-activity relationship studies. Biopolymers 55:74-87. [DOI] [PubMed] [Google Scholar]
- 3.Brissette, C. A., and S. A. Lukehart. 2002. Treponema denticola is resistant to human β-defensins. Infect. Immun. 70:3982-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cociancich, S., A. Ghazi, C. Hetru, J. A. Hoffmann, and L. Letellier. 1993. Insect defensin, an inducible antibacterial peptide, forms voltage-dependent channels in Micrococcus luteus. J. Biol. Chem. 268:19239-19245. [PubMed] [Google Scholar]
- 5.Fujii, G., M. E. Selsted, and D. Eisenberg. 1993. Defensins promote fusion and lysis of negatively charged membranes. Protein Sci. 2:1301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganz, T. 1999. Defensins and host defense. Science 286:420-421. [DOI] [PubMed] [Google Scholar]
- 7.Ganz, T., M. E. Selsted, D. Szklarek, S. S. Harwig, K. Daher, D. F. Bainton, and R. I. Lehrer. 1985. Defensins: natural peptide antibiotics of human neutrophils. J. Clin. Investig. 76:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García, J. R., F. Jaumann, S. Schulz, A. Krause, J. Rodríguez-Jiménez, U. Forssmann, K. Adermann, E. Kluver, C. Vogelmeier, D. Becker, R. Hedrich, W. G. Forssmann, and R. Bals. 2001. Identification of a novel, multifunctional beta-defensin (human beta-defensin 3) with specific antimicrobial activity. Its interaction with plasma membranes of Xenopus oocytes and the induction of macrophage chemoattraction. Cell Tissue Res. 306:257-264. [DOI] [PubMed] [Google Scholar]
- 9.García, J. R., A. Krause, S. Schulz, F. J. Rodríguez-Jiménez, E. Klüver, K. Adermann, U. Forssmann, A. Frimpong-Boateng, R. Bals, and W. G. Forssmann. 2001. Human β-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 15:1819-1821. [PubMed] [Google Scholar]
- 10.Gera, J. F., and A. Lichtenstein. 1991. Human neutrophil peptide defensins induce single strand DNA breaks in target cells. Cell. Immunol. 138:108-120. [DOI] [PubMed] [Google Scholar]
- 11.Gropp, R., M. Frye, T. O. Wagner, and J. Bargon. 1999. Epithelial defensins impair adenoviral infection: implication for adenovirus-mediated gene therapy. Hum. Gene Ther. 10:957-964. [DOI] [PubMed] [Google Scholar]
- 12.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 1997. A peptide antibiotic from human skin. Nature 387:861. [DOI] [PubMed] [Google Scholar]
- 13.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 14.Harwig, S. S., T. Ganz, and R. I. Lehrer. 1994. Neutrophil defensins: purification, characterization, and antimicrobial testing. Methods Enzymol. 236:160-172. [DOI] [PubMed] [Google Scholar]
- 15.Hill, C. P., J. Yee, M. E. Selsted, and D. Eisenberg. 1991. Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization. Science 251:1481-1485. [DOI] [PubMed] [Google Scholar]
- 16.Hoover, D. M., K. R. Rajashankar, R. Blumenthal, A. Puri, J. J. Oppenheim, O. Chertov, and J. Lubkowski. 2000. The structure of human beta-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 275:32911-32918. [DOI] [PubMed] [Google Scholar]
- 17.Hristova, K., M. E. Selsted, and S. H. White. 1996. Interactions of monomeric rabbit neutrophil defensins with bilayers: comparison with dimeric human defensin HNP-2. Biochemistry 35:11888-11894. [DOI] [PubMed] [Google Scholar]
- 18.Hristova, K., M. E. Selsted, and S. H. White. 1997. Critical role of lipid composition in membrane permeabilization by rabbit neutrophil defensins. J. Biol. Chem. 272:24224-24233. [DOI] [PubMed] [Google Scholar]
- 19.Jia, H. P., B. C. Schutte, A. Schudy, R. Linzmeier, J. M. Guthmiller, G. K. Johnson, B. F. Tack, J. P. Mitros, A. Rosenthal, T. Ganz, and P. B. McCray, Jr. 2001. Discovery of new human beta-defensins using a genomics-based approach. Gene 263:211-218. [DOI] [PubMed] [Google Scholar]
- 20.Kagan, B. L., M. E. Selsted, T. Ganz, and R. I. Lehrer. 1990. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc. Natl. Acad. Sci. USA 87:210-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maget-Dana, R., and M. Ptak. 1997. Penetration of the insect defensin A into phospholipid monolayers and formation of defensin A-lipid complexes. Biophys. J. 73:2527-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandal, M., M. V. Jagannadham, and R. Nagaraj. 2002. Antibacterial activities and conformations of bovine beta-defensin BNBD-12 and analogs:structural and disulfide bridge requirements for activity. Peptides 23:413-418. [DOI] [PubMed] [Google Scholar]
- 23.Merlin, D., G. Yue, W. I. Lencer, M. E. Selsted, and J. L. Madara. 2001. Cryptdin-3 induces novel apical conductance(s) in Cl- secretory, including cystic fibrosis, epithelia. Am. J. Physiol. Cell Physiol. 280:C296-C302. [DOI] [PubMed] [Google Scholar]
- 24.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 25.Porter, E. M., E. van Dam, E. V. Valore, and T. Ganz. 1997. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect. Immun. 65:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raj, P. A., and A. R. Dentino. 2002. Current status of defensins and their role in innate and adaptive immunity. FEMS Microbiol. Lett. 206:9-18. [DOI] [PubMed] [Google Scholar]
- 27.Schibli, D. J., H. N. Hunter, V. Aseyev, T. D. Starner, J. M. Wiencek, P. B. McCray, Jr., B. F. Tack, and H. J. Vogel. 2002. The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J. Biol. Chem. 277:8279-8289. [DOI] [PubMed] [Google Scholar]
- 28.Schnölzer, M., P. Alewood, A. Jones, D. Alewood, and S. B. H. Kent. 1992. In situ neutralization in Boc-chemistry solid phase peptide synthesis. J. Peptide Protein Res. 40:180-193. [DOI] [PubMed] [Google Scholar]
- 29.Sharma, S., and G. Khuller. 2001. DNA as the intracellular secondary target for antibacterial action of human neutrophil peptide-I against Mycobacterium tuberculosis H37Ra. Curr. Microbiol. 43:74-76. [DOI] [PubMed] [Google Scholar]
- 30.Starner, T. D., W. E. Swords, M. A. Apicella, and P. B. McCray, Jr. 2002. Susceptibility of nontypeable Haemophilus influenzae to human β-defensins is influenced by lipooligosaccharide acylation. Infect. Immun. 70:5287-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tam, J. P., Y. A. Lu, and Q. Yu. 1999. Thia Zip reaction for synthesis of large cyclic peptides: mechanisms and applications. J. Am. Chem. Soc. 121:4316-4324. [Google Scholar]
- 32.Tang, Y. Q., J. Yuan, G. Osapay, K. Osapay, D. Tran, C. J. Miller, A. J. Ouellette, and M. E. Selsted. 1999. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated α-defensins. Science 286:498-502. [DOI] [PubMed] [Google Scholar]
- 33.Thevissen, K., R. W. Osborn, D. P. Acland, and W. F. Broekaert. 2000. Specific binding sites for an antifungal plant defensin from dahlia (Dahlia merckii) on fungal cells are required for antifungal activity. Mol. Plant-Microbe Interact. 13:54-61. [DOI] [PubMed] [Google Scholar]
- 34.White, S. H., W. C. Wimley, and M. E. Selsted. 1995. Structure, function, and membrane integration of defensins. Curr. Opin. Struct. Biol. 5:521-527. [DOI] [PubMed] [Google Scholar]
- 35.Wimley, W. C., M. E. Selsted, and S. H. White. 1994. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 3:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Wu, Z., D. M. Hoover, D. Yang. C. Boulégue, F. Santamaria, J. J. Oppenheim, J. Lubkowski, and W. Lu. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human β-defensin-3. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 36.Yang, D., O. Chertov, and J. J. Oppenheim. 2001. Participation of mammalian defensins and cathelicidins in anti-microbial immunity: receptors and activities of human defensins and cathelicidin (LL-37). J. Leukoc. Biol. 69:691-697. [PubMed] [Google Scholar]
- 37.Yeaman, M. R., A. S. Bayer, S. P. Koo, W. Foss, and P. M. Sullam. 1998. Platelet microbicidal proteins and neutrophil defensin disrupt the Staphylococcus aureus cytoplasmic membrane by distinct mechanisms of action. J. Clin. Investig. 101:178-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, Q., R. I. Lehrer, and J. P. Tam. 2000. Engineered salt-insensitive alpha-defensins with end-to-end circularized structures. J. Biol. Chem. 275:3943-3949. [DOI] [PubMed] [Google Scholar]