Abstract
Microcin C51 (MccC51) is an antimicrobial nucleotide-heptapeptide produced by a natural Escherichia coli strain. A 5.7-kb fragment of the pC51 plasmid carrying the genes involved in MccC51 production, secretion, and self-immunity was sequenced, and the genes were characterized. The sequence of the MccC51 gene cluster is highly similar to that of the MccC7 gene. Recombinant plasmids carrying different combinations of the mcc genes involved in the MccC51 production or immunity were constructed to characterize their functional roles. The mccA, mccB, mccD, and mccE genes are involved in MccC51 production, while the mccC and mccE genes are responsible for immunity to MccC51. The mcc gene cluster is flanked by 44-bp direct repeats. Amino acid sequence comparisons allowed us to propose functions for each Mcc polypeptide in MccC51 biosynthesis. Plasmid pUHN containing the cloned mccA, mccB, mccC, and mccE genes, but lacking mccD, directed the synthesis of MccC51p, a substance chemically related to MccC51. MccC51p exhibited weak antibiotic activity against E. coli and was toxic to the producing cells. The immunity to exogenous MccC51 determined by the mccC and mccE genes did not overcome the toxic action of MccC51p on the producing cells. The G+C content of the MccC51 operon, markedly lower than that of the E. coli genome, and the presence of direct repeats suggest the possibility of horizontal transfer of this gene cluster.
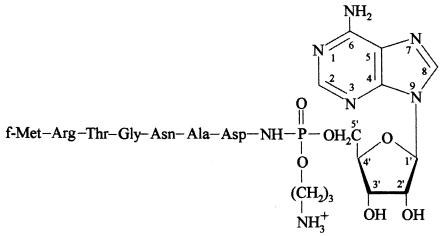
Microcins are gene-encoded antimicrobial peptides secreted by members of the Enterobacteriaceae and are active against closely related bacteria. In most cases, microcin synthesis is controlled by plasmids, which also carry genes responsible for the immunity of the microcin-producing cells (2, 15, 16, 26). They have been recently classified into modified and unmodified microcins (4, 22). Microcin C51 (MccC51) belongs to the modified microcin class. It is a nucleotide-peptide antibiotic (1,178 Da) produced by a natural strain of Escherichia coli isolated in Moscow (Russia) from the feces of a healthy child (14). A recent reinvestigation of the MccC51 structure (3) showed that it differs from the previously published structure (18). It is a heptapeptide with an N-formyl methionine and a C-terminal aspartate linked to AMP aminopropanol through a phosphoramide bond (Fig. 1). The MccC51 structure is identical to that of MccC7 purified from an E. coli strain isolated in Spain (10). MccC51 exhibits a strong antibacterial activity, since it inhibits the growth of different E. coli strains when present at 0.1 to 0.5 μg/ml. The spectrum of MccC51 activity includes various genera and species of Enterobacteriaceae such as Enterobacter, Citrobacter, Klebsiella, Salmonella, Shigella, and Yersinia (14, 17). MccC51 inhibits protein synthesis (I. Khmel et al., unpublished data), as previously shown for MccC7 (8). However, the mechanism of this inhibition remains unresolved.
FIG. 1.
Structure of MccC51.
The genes controlling MccC51 production and immunity have been cloned and mapped. The plasmid genes are clustered within a 5.7-kb DNA fragment and form an operon. Mutation and complementation experiments showed that at least three genes are required for microcin production and two genes are involved in the expression of immunity to the microcin (6, 17).
In the present paper, we describe a detailed analysis of the plasmid genes involved in MccC51 biosynthesis and specify their respective roles. We report the production of MccC51p, an antibiotic substance related to MccC51, by an E. coli strain that harbored the plasmid carrying all the genes of the MccC51 operon except mccD. Nucleotide sequence analysis supports the possibility of a horizontal transfer of this operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The E. coli K-12 strains and plasmids used in this study are listed in Table 1. All strains were grown at 37°C in a minimal medium (13) supplemented with 0.3% yeast extract (Difco), Luria-Bertani (LB) broth, or LB agar (1.5% Ferak). For production of MccC51p, M63 minimal medium (19) was used. Ampicillin and tetracycline were added at final concentrations of 100 and 10 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics and/or genotypea | Source or reference |
|---|---|---|
| E. coli strains | ||
| XL-1 Blue | endA1 gyrA96 hsdR17 lac F′ [proAB+laqIqZΔM15 Tn 10 (Tetr)] recA1 relA1 supE44 thi-1 | Laboratory collection |
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′ [traD36 proAB+lacIqZΔM15] | Laboratory collection |
| Plasmids | ||
| pUC19 | Cloning vector; Apr | 27 |
| pBluescript SK(+) | Cloning vector; Apr | Stratagene |
| pUHAB | pUC19 carrying mccABCDE genes on a 11.2-kb cloned DNA fragment; Apr | 17 |
| pUM5 | pUC19 carrying mccABCDE genes on a 13.5-kb cloned DNA fragment; Apr | 17 |
| pBM43 | pBR325 carrying mccABCDE genes on a 13.5-kb cloned DNA fragment; Apr Cmr | 17 |
| pFUSM | pRS415 carrying the Pmcc-regulatory region and mccA gene (300 bp); Apr | 7 |
| pUHP | pUC19 carrying the Pmcc-regulatory region and mccA gene (300 bp); Apr | This study |
| pSKP | pBluescript SK(+) carrying the Pmcc-regulatory region and mccA gene (300 bp); Apr | This study |
| pUKHn | pUC19 carrying Pmcc and mccACDE genes; Apr | This study |
| pUH1 | pUC19 carrying Pmcc and mccADE genes; Apr | This study |
| pUHA | pUC19 carrying Pmcc and mccABC genes; Apr | This study |
| pUHC | pUC19 carrying Pmcc and mccAC genes; Apr | This study |
| pUHN | pUC19 carrying Pmcc and mccABCE genes; Apr | This study |
| pSKD | pBluescript SK(+) carrying the SacI-SacI(1) fragment of pUM5; Apr | This study |
| pUHE | pUC19 carrying Pmcc and mccAE genes; Apr | This study |
| pUM5-L | pUM5 without the EcoRI-EcoRI fragment containing Pmcc, mccABCD genes, and part of mccE gene; Apr | This study |
| pBM43-L | pBM43 without the EcoRI-EcoRI fragment containing Pmcc, mccABCD genes, part of mccE, and part of pBR325 (EcoRI-BamHI fragment); Apr | This study |
Apr, Cmr, Tetr, resistance to ampicillin, chloramphenicol, and tetracycline, respectively.
Microcin production and immunity assays.
Microcin production by recombinant clones was probed by growing the colonies for 1 to 2 days on minimal medium supplemented with yeast extract and then killing the cells with chloroform vapor. The plates were then overlaid with the MccC51-sensitive E. coli B strain (1 × 107 to 2 × 107 cells in 3 ml of 0.7% agar). Immunity of the different strains to MccC51 was assayed by two methods. The first was derived from the microcin production assay. The strains were considered immune (or partially immune) to MccC51 if the growth inhibition zones surrounding the colonies of the MccC51-producing strain on a lawn of strains to be tested were absent or decreased compared to those observed with the sensitive E. coli K-12 XL-1 Blue strain. In the second assay, 5-μl serial dilutions of MccC51 were spotted on minimal medium supplemented with 0.3% yeast extract plates overlaid with 3 ml of soft agar (0.7%) containing 2 × 107 to 3 × 107 bacteria to be tested for microcin immunity. For both methods, the plates were examined for growth inhibition after overnight incubation.
Plasmid, construction and DNA manipulations.
Plasmid DNA isolation, restriction endonuclease digestion, ligation of DNA fragments with T4 DNA ligase, gel electrophoresis and transformation of competent E. coli cells by plasmid DNA were performed using standard procedures (27). DNA was sequenced by the dideoxynucleotide chain termination method (28). Deletion derivatives of plasmid pUHAB were constructed as follows (Fig. 2). For pUHP, the BamHI-EcoRI fragment of pFUSM carrying the promoter of the MccC51 operon and mccA gene (7) was cloned into pUC19. This fragment was also cloned in pBluescript SK(+), and the resulting recombinant plasmid was named pSKP. For pUKHn, (i) the BglII-BglII fragment of pUHAB was removed, and the plasmid ends were ligated; (ii) the BamHI-KpnI fragment of pSKP was inserted into this construct at BamHI and KpnI sites; and (iii) the BglII-BamHI fragment of this new construct was cut out, and the plasmid ends were ligated. pUH1 was constructed by cutting out the Bsp1407I-Bsp1407I fragment from pUHAB, followed by ligation. pUHA was obtained by cutting out the HindIII(1)-HindIII(2) DNA fragment from pUHAB, followed by ligation. For pUHC, the HindIII(1)-HindIII(2) DNA fragment was cut out from pUKHn, and the ends were ligated. For pUHN, (i) the SacI-SacI(1) fragment of pUM5 was cloned into the SacI site of pBluescript SK(+) so that the SacI(1) site was located near the HindIII site of pBluescript SK(+), and the resulting plasmid was named pSKD; (ii) the HindIII-HindIII fragment of pSKD was cut out and inserted into pUHAB in place of its HindIII(1)-HindIII(2) fragment, leading to pUHN. Thus, pUHN contained all genes of the microcin operon except mccD. For pUHE, the BamHI-KpnI DNA fragment of pSKP was inserted into pSKD and the HindIII(1)-KpnI fragment of the construct obtained was inserted into pUC19. The pUM5-L and pBM43-L plasmids were constructed by cutting out the EcoRI-EcoRI DNA fragment of pUM5 and pBM43, respectively, followed by ligation.
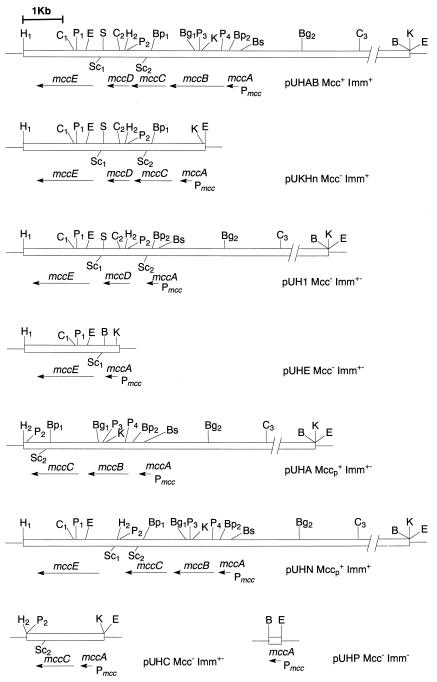
FIG. 2.
Physical and genetic maps of plasmid pUHAB and its deletion derivatives. The white boxes represent DNA fragments from plasmid pC51. The horizontal arrows indicate the direction of transcription. Abbreviations: Mcc+, synthesis of MccC51; Mcc−, absence of MccC51 synthesis; Mccp+, synthesis of MccC51p; Imm+, immunity to microcin; Imm−, absence of immunity to microcin; Imm+−, partial immunity to microcin; P mcc, promoter of the MccC51 operon; mccA, mccB, mccC, mccD, mccE, genes involved in MccC51 production and immunity. Restriction sites: B, BamHI; Bg, BglII; Bp, Bsp1407I; Bs, Bsp119I; C, ClaI; E, EcoRI; H, HindIII; K, KpnI; P, PstI; S, SmaI; Sc, SacI.
The nucleotide and deduced amino acid sequences were compared and analyzed with those available in the Swiss-Prot (release 40.32) and EMBL (release 22.0) databases by using the FASTA (21) or NCBI BLAST 2 (1) program.
Purification of MccC51 and MccC51p.
MccC51 was purified as described previously (3, 18) and further analyzed by reversed-phase high-performance liquid chromatography (RP-HPLC). An antimicrobial substance termed MccC51p was purified from 100-ml culture of E. coli XL-1 Blue harboring the pUHN plasmid. The cells were grown in M63 minimal medium containing 0.2% glucose and 0.3% yeast extract to stationary phase and then centrifuged. The supernatant was applied to a preparative C8 cartridge (BondElut, Varian, France) that was washed with methanol-water (25:75) before the elution of MccC51p with methanol-water (50:50). The crude MccC51p was further purified by RP-HPLC on an Uptisphere C18 column (granulometry, 5 μm; 4.5 by 250 mm; Interchim), with a five-step linear gradient of methanol in water containing 0.05% trifluoroacetic acid. The separation was performed at a flow rate of 1 ml/min, and absorbance was monitored at 220 nm. Active fractions were identified by the growth inhibition assay. The samples were analyzed by electrophoresis at pH 1.5 on Whatman 3MM paper in a formic acid-acetic acid-water mixture (28:20:52) at 270 V for 2.5 h with N-dinitrophenyllysine as the reference standard. The active compounds were localized by a bioassay. Briefly, after being dried for 24 h, the paper was applied to an M63 agar plate for 10 min. The plate was then overlaid with the MccC51-sensitive E. coli B strain. After overnight incubation, detection of growth inhibition zones revealed the location of active compounds.
Nucleotide sequence accession number.
The nucleotide sequence of the DNA fragment containing the MccC51 operon has been deposited in the EMBL database under accession number AJ487788.
RESULTS
MccC51 operon nucleotide sequence.
Plasmid pUHAB, carrying a 11.2-kb cloned DNA fragment of the pC51, contained all the genes required for MccC51 production and immunity (Fig. 2). To study their respective roles, we determined the nucleotide sequence on both DNA strands (5.7 kb). The nucleotide sequence of the MccC51 operon contains five open reading frames (ORF). The first ORF, of 21 nucleotides (mccA), encodes the heptapeptide moiety of MccC51. This ORF is followed by the mccB, mccC, mccD, and mccE ORFs. Downstream of mccE, a 258-bp sequence practically identical to that of the 3′ end of mccF of the MccC7 gene cluster was found.
The G+C content determined for both the MccC51 and MccC7 gene clusters was 34%, a value lower than that found for the E. coli genome (51%). Such a feature was also observed with the G+C content of gene clusters involved in the production of MccB17 (accession no. M24253 and X07875), MccE492 (accession no. AF063590), MccJ25 (accession no. AF061787), MccH47 (accession no. AJ009631 and AJ278866), MccL (accession no. AY237108) and MccM (accession no. AJ515251 and AJ515252). The nucleotide sequence of the MccC51 operon was highly similar to that of MccC7, with a sequence identity varying from 100% for mccA to 98% for mccB, mccC, mccD, and mccE. This result suggested a common origin for the two gene clusters and allowed us to assume that the characteristics of the genes involved in MccC7 production and immunity (9) are associated with the MccC51 genetic system as well. Due to this connection, the genes of the MccC51 operon have been renamed mcc, like those of the MccC7 genetic system. Thus, mccA, mccB, and mccD correspond to those previously termed micD, micC, and micB, respectively, and mccE corresponds to the previously described micA and immA genetic determinants (6, 17).
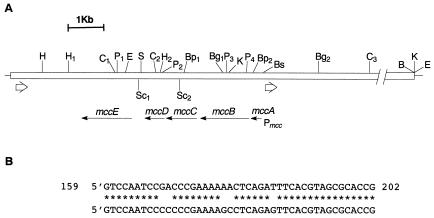
A study of the DNA fragments surrounding the MccC51 operon in plasmid pUM5 that harbored the 13.5-kb cloned fragment of the pC51 plasmid revealed the presence of 44-bp direct repeats flanking the MccC51 gene cluster (Fig. 3A). The repeated sequences (Fig. 3B) were located upstream of mccA (bp 159 to 202) and downstream of mccE (Fig. 3A). The latter sequence exhibited 97.7% identity to the pMccC7 plasmid sequence from bp 6323 to 6366, a region located downstream of mccF in the MccC7 genetic system (9). It is noteworthy that the region encompassing the first repeat sequence has not been sequenced in the case of MccC7. In the MccC51 operon, between the repeat sequence (bp 159 to 202) and mccA (bp 507 to 530), we also found small inverted repeats. Analysis of the sequences next to the direct repeats did not reveal any similarity to insertion sequence elements, transposases, or integrases; the origin of these repeats remains unknown.
FIG. 3.
(A) Location of the direct repeats flanking the genes involved in MccC51 production and immunity. The white box represents part of the DNA fragment from plasmid pC51 contained in plasmid pUM5 (Mcc+, Imm+). The white arrows indicate the direct repeats. The thin horizontal arrows indicate the direction of transcription. For abbreviations, see the legend to Fig. 2. (B) Comparison of the nucleotide sequences of the direct repeats. The upper repeated sequence is located upstream of the microcin operon promoter P mcc, and the lower one is located downstream of mccE.
Roles of the plasmid genes in MccC51 production and immunity.
To study the role of the genes required for MccC51 biosynthesis, deletion constructs of plasmid pUHAB carrying different combinations of genes were generated (Fig. 2) such that the transcription of the cloned genes proceeded from the microcin promoter Pmcc.
From the immunity assay, three patterns were obtained for the different constructs (Fig. 2). The E. coli XL-1 Blue cells carrying no plasmids or plasmid pUHP were sensitive (Imm−) to 0.01 μg of MccC51 per ml. The plasmids carrying both mccC and mccE (pUHAB, pUKHn, and pUHN) determine resistance (Imm+) to >5 μg of MccC51 per ml. Partial immunity (Imm+−) to exogenous microcin was observed with bacterial strains harboring the plasmids carrying either mccC or mccE (pUHC, pUHE, pUH1, and pUHA). The level of immunity in this case was >10 times lower than that of cells carrying both the mccC and mccE genes. Nevertheless, for the cells harboring pUHA, partial immunity was detected only immediately after plasmid construction and bacterial transformation. To determine the presence or the absence of a third MccC51 immunity determinant, we constructed plasmids pUM5-L and pBM43-L carrying the DNA fragment located between mccE and the repeated sequence; this region corresponds to the one carrying mccF in the MccC7 gene cluster (9). The obtained plasmids did not confer any immunity to MccC51 on the cells. It can be hypothesized that mccF is present in the MccC51 gene cluster but that it is defective, presumably due to some frameshift or substitution mutations in the nucleotide sequence.
E. coli cells carrying plasmid pUHP with the single mccA gene did not exhibit any antibiotic activity (Fig. 2). In contrast to the results observed for MccC7 (9), growth of the pUHP plasmid-harboring cells in LB medium was not slowed compared to that of the same bacteria without pUHP. Elimination of mccB from pUHAB abolished MccC51 production. We did not succeed in cloning only mccA and mccB within the same plasmid, presumably as a result of the antibiotic activity exerted by the heptapeptide modified by MccB in the absence of the MccC51 immunity determinants. To verify this hypothesis, we constructed plasmid pUHA carrying the mccA and mccB genes and the immunity determinant mccC. Very small halos of growth inhibition were observed around colonies of TG1 or XL-1 Blue strains that harbored this plasmid, indicating the production of an antibiotic substance. The cells grew slowly and formed smaller colonies than those of cells harboring plasmid pUHAB. After one or two passages, the bacteria lost the ability to produce the antibiotic, due to the selection of mutants unable to synthesize it. In addition, the cells harboring plasmid pUHA became susceptible to exogenous MccC51. This result could explain the unexpected Imm− phenotypes of both plasmid pUHA (17) and plasmid pMM554, which carries the same DNA fragment in the MccC7 gene cluster (20). Thus, the presence in plasmid pUHA of mccC was insufficient to protect E. coli cells against the antibiotic substance. Therefore, plasmid pUHN, which also carries the mccE immunity gene, was constructed. It endowed the producing cells with immunity to MccC51. As also observed for bacteria harboring pUHA, small halos of growth inhibition around colonies of E. coli XL-1 Blue harboring pUHN suggested the production of an antibiotic substance that we termed MccC51p. Although plasmid pUHN carried the mccC and mccE immunity genes, the bacteria harboring this plasmid very quickly lost their ability to produce the antibiotic substance and grew poorly, presumably because of a toxic effect of MccC51p on the cells that produced it.
Amino acid sequence comparison of the deduced mcc gene products.
Considering the high amino acid sequence identity between the deduced mcc gene products from the MccC51 and MccC7 gene clusters (100% for MccA and 97.7% and 8 amino acid substitutions over 350 residues for MccB), the features observed previously for MccC7 (9) may apply to the polypeptides from the MccC51 operon. Nevertheless, comparisons with the current releases of SwissProt and EMBL databases revealed new characteristics, especially for MccD and the N-terminal region of MccE, for which significant homologies were not detected in the earlier study (9). The relevant sequence homologies of Mcc proteins are summarized in Table 2.
TABLE 2.
Gene products of the MccC51 operon and homologies to proteins in the databases
| Location in nucleotide sequence | Gene product (size)a | Homology tob: | Origin | Identity (%)/similarity (%) (range)c | Amino acid sequence accession no. |
|---|---|---|---|---|---|
| 605-1657 | MccB (350 aa) | MccB-like protein | H. pylori | 30/48 (27-348) | Q8L2W7 |
| ThiF/MoeB/HesA proteins | Various bacteria | 24-28/43-49 (110-347) | P12282d | ||
| P30138d | |||||
| 1654-2868 | MccC (404 aa) | Multidrug efflux transporter | Borrelia burgdorferi | 21/41 (29-389) | O50880 |
| Multidrug resistance protein | Fusobacterium nucleatum | 21/37 (27-397) | Q8RIS2 | ||
| Hypothetical protein | H. pylori | 21/43 (29-395) | Q9F7U3 | ||
| 2865-3668 | MccD (267 aa) | Menaquinone biosynthesis methyl- transferase | Staphyloccus aureus | 35/52 (48-149) | Q99U19 |
| UbiE | Pasteurella multocida | 31/48 (48-180) | Q9CKD6 | ||
| Methyltransferase | Streptococcus agalactiae | 25/45 (22-191) | Q9F8B8 | ||
| 3856-5421 | MccE (521 aa) | Diaminopimelate decarboxylase | Bacillus methanolicus | 26/42 (1-312) | P41023 |
| LysA protein | Listeria monocytogenes | 23/41 (3-312) | Q8Y5V3 | ||
| RimL | E. coli | 41/62 (349-520) | P13857 | ||
| RimJ | Rickettsia prowazekii | 36/53 (416-520) | Q9ZCN0 |
aa, amino acid.
Thi; thiamine; Moe; molybdopterin; Ubi; ubiquinone.
Contiguous stretch from which the identity and similarity were calculated.
Chosen among numerous proteins.
MccB displayed significant homology to both an MccB-like protein and a hypothetical protein from Helicobacter pylori. Furthermore, as previously observed (9), MccB exhibited homology to known or putative bacterial proteins involved in the adenylation of the carboxyl-terminal glycine of thiamine synthetase and of molybdopterin synthase. All these proteins, like MccB, contain a nucleotide-binding domain. MccC showed a low but significant degree of homology to the multidrug efflux transporters as well as to two putative proteins from H. pylori. MccD had significant homology to bacterial proteins of the methyltransferase family. Amino acid sequence comparisons strongly indicated that MccE contains two putative domains. Its N-terminal sequence up to the Arg-312 residue was similar to sequences of pyridoxal phosphate-dependent diaminopimelic acid decarboxylase from several species of Bacillus, Listeria, and other bacteria. On the other hand, the C-terminal region of MccE is highly homologous to RimL from E. coli and Salmonella and to RimJ from Rickettsia conorii and R. prowazekii, two proteins that catalyze the acetylation of N-terminal alanine or serine residues of ribosomal proteins (29, 31).
Despite the absence of a functional mccF gene in the MccC51 operon, we reinvestigated the amino acid sequence homology of the MccF polypeptide (344 residues) deduced from the MccC7 genetic system (9). The highest score was obtained with the hypothetical protein VCA0337 from Vibrio cholerae, with 56% identity and no gaps in the alignment. Other significant homology (27 to 40% identity) was observed to the l,d-carboxypeptidase A from E. coli (accession no. P76008) and uncharacterized proteins from various bacterial genera (such as Bacillus, Clostridium, Listeria, and Streptococcus).
Isolation and partial characterization of MccC51p.
Only a small amount of MccC51p was obtained from the culture supernatant of the strain harboring pUHN, so that we were unable to perform a structural analysis. The antibiotic activity of MccC51p was about 50 to 100 times weaker than that of MccC51. RP-HPLC analyses showed MccC51p to be more hydrophobic than the mature MccC51, since MccC51 and MccC51p were eluted with 22 and 34% methanol, respectively. The similarity of the MccC51p and MccC51 UV absorption spectra, both showing two maxima at 260 and 205 nm, suggested that both MccC51 and MccC51p included the peptide and the nucleotide moieties. The electrophoretic mobility of MccC51p was approximately 65% of that of MccC51. Under the experimental conditions used, the mobility of peptides is proportional to the number of ionizable amino groups. Since MccC51 contains three free amino groups, one can hypothesize that MccC51p bears only two amino groups, which could explain its higher hydrophobicity. Since the cells harboring pUHN synthesized the heptapeptide moiety encoded by the mccA gene, this result suggests that MccC51p contains only one amino group in its C-terminal substituent.
DISCUSSION
The genetic system and the roles of the genes required for MccC51 production and immunity were analyzed in detail in the present study. The amino acid sequence homologies associated with the expression of various constructs carrying one to four plasmid genes of the MccC51 operon allowed us to ascribe roles to the deduced mcc gene products. The gene cluster determining MccC51 production, secretion, and immunity contains five genes, mccA, mccB, mccC, mccD, and mccE. mccA, mccB, mccD, and mccE are involved in MccC51 production, and mccE also contributes to microcin immunity, along with mccC. The nucleotide sequences of these genes are very similar to those of the genes involved in MccC7 production and immunity. In practice, the same genes participate in microcin biosynthesis in both producing strains isolated in geographically distant regions.
The plasmids carrying either mccC or mccE did not confer complete immunity to the microcin-producing bacteria. Complete immunity was obtained only with constructs harboring both mccC and mccE. The amino acid sequence homology of MccC and the multidrug efflux transporter is consistent with both its location in the bacterial membrane, as previously suggested for MccC7 (9), and an involvement in the export of MccC51 outside the cells. The homology of the C-terminal region of MccE to RimL and RimJ, enzymes known to acetylate the N-terminal residue of ribosomal proteins (29, 31), suggests that MccE could act by acetylating the intracellular target of microcin on the ribosome, making it insensitive to the antibiotic. This proposition is consistent with the effect of MccC51 (Khmel et al., unpublished) and MccC7 (8, 10) on protein synthesis. In addition to mccC and mccE, the mccF gene, located downstream of mccE, is involved in MccC7 immunity (9). We did not detect a third immunity determinant in the MccC51 gene cluster when cloning the same region from plasmids pUM5 and pBM43. Thus, only the mccC and mccE genes determine immunity to MccC51, and mccF is not involved in this immunity, differing on that point from the situation for MccC7. The mccF gene is transcribed in the opposite direction from the other genes and contributes weakly to the complete immunity to MccC7 (9). On the other hand, the similarity of MccF to E. coli l,d-carboxypeptidase A, an enzyme involved in the turnover and recycling of peptidoglycan (31), and to numerous hypothetical proteins encountered in various bacterial species, not known to produce microcins, would indicate that MccF protein is not restricted to MccC7 immunity and appears to exhibit another function widely distributed throughout bacterial species.
The amino acid sequence homology of MccB to nucleotide-binding proteins involved in adenylation processes strongly suggests that this protein contributes to the substitution of the C terminus of the heptapeptide by AMP. The absence of the mccD gene in pUHN carrying all the other genes of the MccC51 operon results in the synthesis of MccC51p, a substance chemically related to MccC51 and toxic for the producing cells. The presence of mccC and mccE is insufficient to protect the bacteria from MccC51p activity. Thus, only the mature MccC51 appears to interact with the immunity proteins determined by mccC and mccE. The electrophoretic mobility indicates that MccC51p has one amino group less than MccC51. This result, together with the similarity between MccD and transferases, suggests that MccD could be involved in the n-aminopropanol transfer to the AMP group. The N-terminal amino acid sequence of MccE shows a conserved domain of ornithine, diaminopimelic acid, and arginine decarboxylases. We have demonstrated previously, by performing transposon Tn 5 insertions in mccE, that MccE can participate in microcin production (17). The amino acid sequence comparison reveals that MccE contains two independent domains and consequently suggests its possible involvement in both MccC51 immunity and production. However, the precise role of MccE in microcin production is still unresolved. Nevertheless, similarly to the ethanolamine synthesis, which can be achieved by direct decarboxylation of serine (5, 25), we can hypothesize that MccE would be responsible for the MccC51 aminopropanol group formation through homoserine decarboxylation.
Analysis of the nucleotide sequence of the MccC51 operon shows for the first time the presence of direct-repeat sequences flanking the cluster of genes involved in microcin biosynthesis. Such repeats might surround the MccC7 gene cluster, as indicated by the sequence homology of the repeat located at the 3′ end of both genetic systems. The G+C contents of both MccC51 and MccC7 gene clusters differ markedly from that of the E. coli chromosome. Therefore, E. coli would appear not to be the original host of microcin genes. The identification of direct repeats flanking the microcin operon and the G+C contents of the gene clusters suggest the possibility of a horizontal transfer of plasmid genes responsible for the MccC51 biosynthesis. Such properties are also characteristic of particular regions of the prokaryotic genomes, termed pathogenicity islands, which are present in various pathogenic species but absent from nonpathogenic bacteria of the same or related species (11). Interestingly, for plasmids pHPM8 and pHel4 carried by H. pylori, the amino acid sequences of two proteins were homologous to those of the MccB and MccC proteins. Furthermore, possible microcin structural genes were also identified upstream of mccB-like genes (12, 23). The presence of part of the MccC51-MccC7 genetic system in plasmids of a bacterium that does not belong to the family Enterobacteriaceae is consistent with an acquisition of microcin gene clusters by horizontal gene transfer. Microcin biosynthesis might provide a selective advantage for the microcin-producing bacteria over nonmicrocinogenic strains, a situation also observed for bacteriocins (24). In that case, the genes involved in microcin production and immunity might participate in the competitions between bacteria in microbial ecosystems. Consequently, selective pressure might lead to the dissemination of the microcin gene cluster among bacterial populations and to its transfer to different species and genera of bacteria.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baquero, F., and F. Moreno. 1984. The microcins. FEMS Microbiol. Lett. 23:117-124. [Google Scholar]
- 3.Blond, A., C. Goulard, D. E. Fomenko, A. Z. Metlitskaya, J. Péduzzi, M. Barthélémy, G. Katrukha, I. Khmel, and S. Rebuffat. 2000. Structure/activity relationship of the antibiotic nucleotide-peptide microcin C51, p. 601-602. In J. Martinez, and J.-A. Fehrentz (ed.), Peptides 2000. Proceedings of the 26th European Peptide Symposium. EDK, Paris, France.
- 4.Destoumieux-Garzón, D., J. Peduzzi, and S. Rebuffat. 2002. Focus on modified microcins: structural features and mechanism of action. Biochimie 84:511-519. [DOI] [PubMed] [Google Scholar]
- 5.Elabbadi, N., M.L. Ancelin, and H.J. Vial. 1997. Phospholipid metabolism of serine in Plasmodium-infected erythrocytes involves phosphatidylserine and direct serine decarboxylation. Biochem. J. 324:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fomenko, D. E., E. I. Basyuk, V. M. Bezrukov, A. A. Volodin, A. Z. Metlitskaya, and I. A. Khmel. 1996. Identification and expression of plasmid genes involved in synthesis of microcin C51. Russ. J. Genet. 32:1149-1155. (Erratum, 33:116, 1997.) [PubMed] [Google Scholar]
- 7.Fomenko, D., A. Veselovskii, and I. Khmel. 2001. Regulation of microcin C51 operon expression: the role of global regulators of transcription. Res. Microbiol. 152:469-479. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Bustos, J. F., N. Pezzi, and E. Méndez. 1985. Structure and mode of action of microcin 7, an antibacterial peptide produced by Escherichia coli. Antimicrob. Agents Chemother. 27:791-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González-Pastor, J. E., J. L. San Millán, M. A. Castilla, and F. Moreno. 1995. Structure and organization of plasmid genes required to produce translation inhibitor microcin C7. J. Bacteriol. 177:7131-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guijarro, J. I., J. E. González-Pastor, F. Baleux, J. L. San Millán, M. A. Castilla, M. Rico, F. Moreno, and M. Delepierre. 1995. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J. Biol. Chem. 270:23520-23532. [DOI] [PubMed] [Google Scholar]
- 11.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 12.Hofreuter, D., and R. Haas. 2002. Characterization of two cryptic Helicobacter pylori plasmids: a putative source of horizontal gene transfer and gene shuffling. J. Bacteriol. 184:2755-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khmel, I. A., V. M. Kopylov, I. P. Vorobjeva, and V. P. Polyanin. 1981. The influence of colicinogenic plasmids ColIb-P9, ColIa-CA53 and ColV-K30 on the repair, mutagenesis and induction of colicin E1 synthesis. Mol. Gen. Genet. 181:101-106. [DOI] [PubMed] [Google Scholar]
- 14.Khmel, I. A., V.M. Bondarenko, I. M. Manokhina, E. I. Basyuk, A. Z. Metlitskaya, V. A. Lipasova, and Y. M. Romanova. 1993. Isolation and characterization of Escherichia coli strains producing microcins of B and C types. FEMS Microbiol. Lett. 111:269-274. [DOI] [PubMed] [Google Scholar]
- 15.Khmel, I. A. 1999. Microcins, peptide antibiotics of enterobacteria: genetic control of synthesis, structure, and mode of action. Russ. J. Genet. 35:1-10. [PubMed] [Google Scholar]
- 16.Kolter, R., and F. Moreno. 1992. Genetics of ribosomally synthesized peptide antibiotics. Annu. Rev. Microbiol. 46:141-146. [DOI] [PubMed] [Google Scholar]
- 17.Kurepina, N. E., E. I. Basyuk, A. Z. Metlitskaya, D. A. Zaitsev, and I. A. Khmel. 1993. Cloning and mapping of the genetic determinants for microcin C51 production and immunity. Mol. Gen. Genet. 241:700-706. [DOI] [PubMed] [Google Scholar]
- 18.Metlitskaya, A. Z., G. S. Katrukha, A. S. Shashkov, D. A. Zaitsev, Ts. A. Egorov, and I. A. Khmel. 1995. Structure of microcin C51, a new antibiotic with a broad spectrum of activity. FEBS Lett. 357:235-238. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Novoa, M. A., L. Diaz-Guerra, J. L. San Millan, and F. Moreno. 1986. Cloning and mapping of the genetic determinants for microcin C7 production and immunity. J. Bacteriol. 168:1384-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pons, A.-M., I. Lanneluc, G. Cottenceau, and S. Sable. 2002. New developments in post-translationnaly modified microcins. Biochimie 84:531-537. [DOI] [PubMed] [Google Scholar]
- 23.Quiñones, M., J. E. Knesek, and S. A. McIntire. 2001. Sequence and gene expression analyses of plasmid pHPM8 from Helicobacter pylori reveal the presence of two operons with putative roles in plasmid replication and antibiotic activity. Plasmid 46:223-228. [DOI] [PubMed] [Google Scholar]
- 24.Riley, M. A., and D. M. Gordon. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7:129-133. [DOI] [PubMed] [Google Scholar]
- 25.Rontein, D., I. Nishida, G. Tashiro, K. Yoshioka, W.I. Wu, D.R. Voelker, G. Basset, and A. D. Hanson. 2001. Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J. Biol. Chem. 276:35523-35529. [DOI] [PubMed] [Google Scholar]
- 26.Salomon, R. A., and R. Farias. 1992. Microcin 25: a novel antimicrobial peptide produced by Escherichia coli. J. Bacteriol. 174:7428-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sanger, F., S. Nicklen, and A.R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka, S., Y. Matsushita, A. Yoshikawa, and K. Isono. 1989. Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol. Gen. Genet. 217:289-293. [DOI] [PubMed] [Google Scholar]
- 30.Templin, M. F., A. Ursinus, and J. V. Holtje. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 18:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshikawa, A., S. Isono, A. Sheback, and K. Isono. 1987. Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen. Genet. 209:481-488. [DOI] [PubMed] [Google Scholar]