Abstract
Among insects, the genetic regulation of regional identities in the postoral head or gnathal segments (mandibular, maxillary, and labial) is best understood in the fly Drosophila melanogaster. In part, normal gnathal development depends on Deformed (Dfd) and Sex combs reduced (Scr), genes in the split Drosophila homeotic complex. The gnathal segments of Dfd and Scr mutant larvae are abnormal but not homeotically transformed. In the red flour beetle, Tribolium castaneum, we have isolated loss-of-function mutations of the Deformed ortholog. Mutant larvae display a strong transformation of mandibular appendages to antennae. The maxillary appendages, normally composed of an endite and a telopodite, develop only the telopodite in mutant larvae. We previously reported that mutations in the beetle Scr and Antennapedia orthologs cause the labial and thoracic appendages, respectively, to be transformed to antennae. Moreover, a deficiency of most of the beetle homeotic complex causes all gnathal (as well as thoracic and abdominal) segments to develop antennae. These and other observations are consistent with the hypothesis that ancestral insect homeotic gene functions have been modified considerably during the evolution of the highly specialized maggot head. One of the ancestral homeobox genes that arose close to the root of the Eumetazoa appears to have given rise to Dfd, Scr, and the Antennapedia homeobox-class homeotic genes. Evidence from both Tribolium and Drosophila suggests that this ancestral gene served to repress anterior development as well as confer a trunk-specific identity.
Scientists were originally attracted to the study of Drosophila homeotic genes by striking adult mutant phenotypes in which one body region is developmentally replaced by another. We now understand that these genes specify region-specific developmental pathways by encoding homeodomain-bearing transcription factors that control the expression of downstream target genes. In his classic studies of the bithorax complex (BXC), Lewis (1) emphasized that BXC functions are necessary to confer progressively more posterior identities in the metathorax (T3) and abdomen. In the absence of BXC activity, these regions display reiterations of a developmental “ground state” later identified as parasegment (PS)4 (posterior first thoracic segment/anterior second thoracic segment, or T1p/T2a) (2). The identity of PS4 in turn depends on the expression of homeotic genes in the Antennapedia complex. If Sex combs reduced (Scr) and Antennapedia (Antp) functions are eliminated in addition to those of the BXC, the reiterated unit includes an abnormal T1a and ill-defined head structures (3, 4).
For technical reasons, no one has examined the development of a Drosophila embryo lacking all homeotic complex (HOMC) functions. One HOMC gene, proboscipedia (pb), is completely dispensable for normal embryogenesis. Normal development of the mandible requires Deformed (Dfd) and cap “n” collar (cnc) (a gene not located in the homeotic complex), whereas Dfd is important for the maxilla and Scr for the labium. Mutations in Dfd and Scr do not result in overt homeotic transformations of larval gnathal segments, which retain a gnathal character but display a loss of segment-specific features (5).
During early embryogenesis, Drosophila has a normal complement of head segments, albeit somewhat reorganized with respect to the linear arrangement typical of most insects (6). However, these segments involute through the stomodeum to elaborate internal structures, and only vestigial portions of the gnathal segments remain external. It is likely that the evolution of the maggot head was accompanied by changes in the genetic control of developmental commitments.
We have been characterizing the HOMC of the red flour beetle, an insect that allows the possibility of sophisticated genetic analysis. Tribolium larvae display relatively unspecialized heads with mandibulate mouth parts. Tribolium has a Deformed ortholog that is located in the HOMC (7) and expressed in an embryonic pattern essentially identical to that in Drosophila (8). We will refer to this beetle gene as Tc Deformed (TcDfd). Here we describe the isolation and mutant phenotypes of TcDfd variants and discuss their implications for the evolution of Hox gene function.
Materials and Methods
Genetics.
The genetic variants used in this study are summarized in Table 1.
Table 1.
Alleles described in this work
| Allele name | Symbol | Description | Reference |
|---|---|---|---|
| maxillopediaStuboid | mxpStbd | Dominant; short antennae; recessive lethal; crossover suppressor | 10 |
| maxillopediaStumpy | mcpStm | Dominant; fused antennae; recessive lethal; crossover suppressor | 10 |
| Cephalothorax5 | Cx5 | Haploinsufficient; T1→T2; induced on a mxpStm chromosome recessive lethal; crossover suppressor | 10 |
| Eyeless | Ey | Dominant; eye facets missing; recessive lethal; crossover suppressor | 10 |
| AbdominalExtra Sclerite | AEs | Dominant; A2→A3; recessive lethal; crossover suppressor | 10 |
| Deficiency (HOMC) | Df (HOMC) | Homeotic genes TcDfd, Cx, ptl, Utx, and A deleted; recessive lethal; MN-A8→AN | 7 |
| tar | tar | Recessive; prothoracic quinone glands darkened | 9 |
| Antennagalia | Ag | Dominant; maxillary galea on base of antennae; recessive lethal; gamma irradiation induced | 7 |
| AgPinhead | AgPin | Dominant; reduced head capsule, tightly linked to Ag; recessive lethal, gamma irradiation induced | This work |
| TcDeformed1 | TcDfd1 | Recessive lethal; EMS induced | This work |
| TcDeformed2 | TcDfd2 | Gamma irradiation induced revertant of AgPin, recessive lethal | This work |
The allele name and symbol are listed. The dominant or haploinsufficient phenotype associated with each allele is listed first. The recessive phenotype is listed second. The first five alleles are dominant markers on balancer chromosomes.
Mutagenesis.
Males homozygous for tar, a recessive viable mutation tightly linked to the HOMC (9), were mutagenized with 0.04 M ethyl methanesulfonate as previously described (10). The tar chromosome was recently isogenized by using a balancer in the region of the HOMC and was shown to be lethal free. Mutagenized males were mated (1:2) with AEs/Ey females. After 3 days, the males were removed, and females were transferred weekly to fresh flour. Individual Es progeny were mated to mxpStbd/Df(HOMC) beetles and their progeny screened for non-Es non-Stbd animals. The absence of this class indicated the presence of a lethal mutation in the region uncovered by the deficiency. Stocks of putative mutants were established by intermating the Es non-Stbd siblings of the absent class. Eight mutants were recovered in a screen of 1,600 F1 individuals, one of which (TcDfd1) is described here.
Reversion.
AgPin/mxpStm Cx5 males were subjected to γ irradiation (6,000 rads), allowed to recover for 2 days, and mated to wild-type females. The males were removed after 4 days and the F1 progeny visually screened for revertants. Putative AgPin revertants were mated to AEs/Ey beetles to allow secondary mutations to segregate and to establish balanced stocks. Four revertants of the AgPin phenotype were recovered in a screen of 9,250 F1 individuals, one of which (TcDfd2) is described here.
Complementation.
Balanced stocks of TcDfd1, TcDfd2, and Df(HOMC) were crossed inter se to assess complementation with respect to viability. The two new variants were also tested and shown to complement mutations at all of the other homeotic genes in the HOMC for which variants are available (data not shown).
Molecular Analysis of TcDfd1.
Primers were designed to amplify the first exon of TcDfd (Fig. 1a). The fragment amplified from a single homozygous mutant larva was gel purified and sequenced with internal primers.
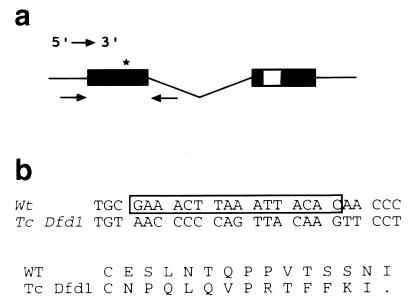
Figure 1.
Molecular analysis of TcDfd1. (a) Diagram of the TcDfd transcription unit. Coding regions are shown as solid boxes, whereas the homeobox in the second exon is hatched. The arrows below exon one denote the primers used to amplify that region. The asterisk marks the location of the molecular lesion. (b) Comparison of wild-type (wt) and TcDfd1 DNA and amino acid sequences. The 16 bases deleted in TcDfd1 are boxed. The shift in DNA reading frame results in a protein truncated after the next 13 residues.
Microscopy.
Larval cuticles were prepared and documented as previously described (11). In situ hybridization to TcDfd mRNA and immunohistochemical analysis were performed as previously described (12). The anti-Drosophila Dll antibody was the kind gift of G. Panganiban (University of Wisconsin–Madison, Madison, WI).
Results
We had previously molecularly cloned and characterized TcDfd and determined that the gene lies within a region of the Tribolium HOMC deleted by Df(HOMC) (7). We have isolated two new mutations at the TcDfd locus. One, TcDfd1, was induced by ethylmethanesulfonate and isolated by its failure to complement Df(HOMC) for viability. The second, TcDfd2, was isolated as a γ ray-induced revertant of the dominant variant Antennagalea-pinhead (Agpin). The original Ag lesion causes a galea (a maxillary structure) to appear on the base of the antenna (7), whereas the derivative pinhead variant additionally causes a reduction of the head capsule. TcDfd1 gives an identical lethal mutant phenotype (see below) in homozygous and hemizygous condition. This phenotype is shared by TcDfd1/TcDfd2 and TcDfd2/Df(HOMC) individuals, suggesting that TcDfd1 and TcDfd2 are loss-of-function (probably null) mutations at the gene. Although Agpin/Df(HOMC) larvae are normal, Agpin homozygotes die as early embryonic lethals. Moreover, the Agpin derivative TcDfd2 gives early embryonic lethality when homozygous and when heterozygous with Agpin. Because Agpin is associated with crossover suppression, it is likely that it and TcDfd2 share a recessive lethal breakpoint outside of the limits of the deficiency. These data suggest Agpin is a gain-of-function variant that retains normal TcDfd function, and that the TcDfd2 revertant was generated by inactivation of the gene.
We have demonstrated that TcDfd1 is associated with a lesion at the molecularly defined TcDfd gene. Genomic DNA from a TcDfd1 homozygous larva was used as a template to amplify the coding portion of the 5′ exon and a short portion of the adjacent intron of the TcDfd gene by using PCR (Fig. 1a). Sequencing revealed that, compared with wild type, there is a C→T transition and an adjacent 16-bp deletion (Fig. 1b). These changes result in a translational frameshift and, 13 codons downstream, a translational stop N-terminal to the homeodomain. This result further supports the likelihood that TcDfd1 is a null mutation. In situ hybridization of wild-type embryos with a probe from TcDfd reveals that the gene is transcribed in the mandibular and maxillary segments (Fig. 2a) (8). When TcDfd1 homozygous embryos are treated similarly, they display a dramatic reduction in signal intensity (Fig. 2b), suggesting that the deletion results in reduced transcript stability. Finally, the apparent TcDfd null phenotype was phenocopied by RNA interference experiments in which double-stranded RNA molecules complementary to a TcDfd cDNA were injected into young wild-type embryos (11).
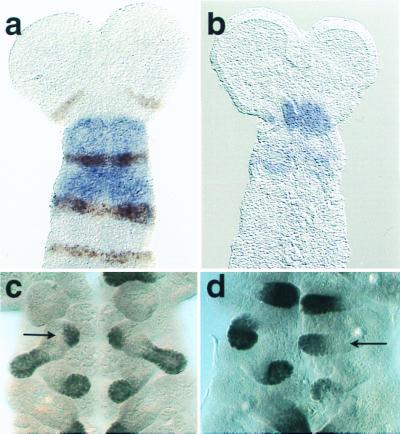
Figure 2.
TcDfd and Dll expression in wild-type and TcDfd1 mutant embryos. (a) TcDfd is expressed throughout the mandibular (arrow) and maxillary segments of wild-type embryos. Engrailed (En) expression marks the posterior compartment of each segment. [Reproduced with permission from ref. 8 (Copyright 1999, Springer)]. (b) TcDfd expression in mutant embryos is severely reduced. (c) Dll is expressed in each limb tip of wild-type embryos except the mandibular. In addition, it is expressed in the developing endite (arrow) of the maxillary appendage. (d) In mutants, Dll is expressed in the transformed mandibular appendage. However, expression normally associated with the developing maxillary endite is missing, and no endite forms.
Fig. 3a shows a ventral view of a wild-type first instar Tribolium larva. The maxillary appendage differs from those of the mandibular and labial segments in having two lobes: a slightly larger distal branch (telopodite) and a ventrally oriented branch (endite) (see Fig. 3 a and c). The labial appendages have only a telopodite that closely resembles those of the maxillary segment, albeit somewhat smaller. Although the labial appendage primordia are initially more widely separated, they move together at the ventral midline and somewhat anteriorly such that they are nested between the maxillary appendages in the first instar larva. The mandibular appendages are stout toothed structures that are gnathobasic: they represent only a proximal coxopodite derived from the body wall (13). As such, they resemble the mandibular primordia of Drosophila and other arthropods in lacking Distal-less (Dll) expression (Fig. 2c) typical of the more distal telopodite of most arthropod appendages (14, 15). During appendage development in the maxillary segment, Dll is expressed in the primordia of both the endite and telopodite, whereas there is a single domain of Dll expression associated with each labial appendage (Fig. 2c).
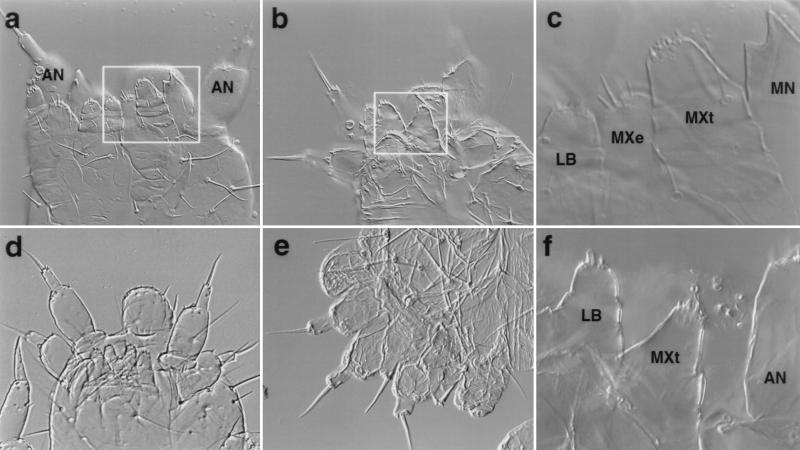
Figure 3.
Wild-type and mutant first instar larval cuticles. (a) Ventral view of a wild-type cuticle. (b) Ventral view of TcDfd1 homozygote. The terminal seta of the homeotic antenna on the right is missing. (c) The boxed region in a photographed at higher magnification. (d) Ventral view of TcDfd1/Df(HOMC). The homeotic antennae on the mandibular segment are fully transformed. (e) Lateral view of TcDfd2/Df(HOMC). Note both normal and homoetic antennae. (f) The boxed region in b photographed at higher magnification. The maxillary endite is missing, and the maxillary telopodite is reduced in size. The homeotic antenna is out of the plane of focus. AN, antennal; LB, labium; MXe, maxillary endite; MXt, maxillary telopodite; MN, mandible; LR, labrum. Lower magnification = ×200; higher magnification = ×400.
In Tribolium, individuals homozygous (Fig. 3b) or hemizygous (Fig. 3 d and e) for TcDfd mutations display a strong transformation of the mandibular appendages to antennae. This transformation is associated with Dll expression in the homeotic mandibular appendages (Fig. 2d). In addition, the maxillary appendages lack endites (Fig. 3f), a change associated with the loss of the normal ventral domain of Dll expression (Fig. 2d). The remaining maxillary telopodites are somewhat smaller, but otherwise appear unchanged. Given the similarity of the maxillary and labial telopodites, it is conceivable that the mutant maxillary appendages have a labial identity. Two observations suggest to us that the identity of the mutant maxillary telopodites is unchanged. First, in normal embryos the developing labial telopodites express both the pb and Scr orthologs, whereas the maxillary telopodites express the pb and Dfd orthologs. Expression of the Scr ortholog is not significantly altered in TcDfd mutants (unpublished observations). Second, in TcDfd mutants the maxillary telopodites do not migrate to the midline as is typical of the labium.
Discussion
Deformed Function in Beetles and Flies.
The Deformed gene of Drosophila plays a role during embryogenesis in specifying maxillary identity, but loss of Dfd function does not result in overt homeotic transformation of that segment. Some ventral structures of maxillary origin (such as the mouth hooks, ventral organs, and some cirri) are missing and thus are Dfd dependent, whereas the features still present are Dfd independent (16). Mouth hooks, ventral organs, and cirri appear in the labial and thoracic segments because of ectopic Dfd expression, confirming the conclusion that they are Dfd-dependent structures. There are two domains of Dll expression in the embryonic maxillary segment: a ventral–lateral domain and a dorsal domain. These also represent Dfd-dependent and -independent features, respectively, and ventral–lateral Dll expression is missing in Dfd mutants (17). Moreover, ventral–lateral Dll expression is necessary for the development of most cirri.
Although Dfd is expressed broadly in the Drosophila mandibular segment, it is not clear that it is important to the establishment of mandibular identity. Dfd mutant larvae are subject to disruptions of the organization of the mandible as well as other head segments but appear to have a full complement of mandibular structures except for a sensory papilla (16). McGinnis et al. (18) have shown that a protein isoform encoded by the cap “n” collar gene selectively interferes with the activation of Dfd response elements in the mandibular segment. In cnc mutants, homeotic maxillary structures develop in the mandibular segment. The genetic functions acting to promote mandibular identity in Drosophila are not entirely clear.
We report here the isolation of two mutations of the Tribolium Deformed ortholog TcDfd. Their phenotypes in hemizygous vs. homozygous condition and the demonstration that one of them encodes a protein that is truncated N-terminal to the homeodomain indicate that they are both nulls or near nulls. Loss of TcDfd function results in a strong homeotic transformation of the mandibular appendages to antennae. Clearly, unlike in flies, TcDfd is important for repressing anterior development in the mandibular segment. It is not clear whether TcDfd also acts to promote mandibular identity in that segment. The role of TcDfd in the beetle maxilla appears to parallel that of its fly ortholog. Beetles resemble flies in having two domains of embryonic maxillary Distal-less expression: one dorsal and one ventral. In each insect, dorsal expression is Dfd-independent, whereas ventral expression is Dfd dependent. Further, mutation in each insect results in the loss of structures in the ventral region (the endite in the case of Tribolium). Jürgens et al. (6) suggested that the cirri, ventral organ, and mouth hooks are all homologous to structures (lacinia or galea) forming parts of the maxillary endite of some other insects. It appears that the Dfd orthologs of both insects act to promote ventral maxillary identity. The observation that Agpin, an apparent TcDfd gain-of-function mutation, causes ectopic appearance of the galea in adults further supports the conclusion that the gene promotes ventral maxillary identity in beetles, as it does in flies.
Evolving Roles of the Antennapedia Complex (ANTC) Genes.
The Drosophila ANTC includes five homeotic selector genes. Probable null mutant phenotypes have now been identified for Tribolium orthologs of four of these genes [to date no Tc labial (Tclab) variant has been isolated]. In each case, the beetle larval mutant phenotype differs significantly from that described in flies (ref. 19 and this work). Most significantly, mutants of the Antp, Scr, and Dfd orthologs include transformations of thoracic, labial, and mandibular appendages, respectively, to antennae [it is likely that the ancestral pb gene function became highly derived before the origin of the insects (5)]. Moreover, in homozygous condition a deficiency of most of the complex results in an antennal transformation of all gnathal, thoracic, and abdominal segments (7). Given that anterior development in Tribolium embryos is far less derived than that in Drosophila, it is likely that the functions of the beetle homeotic genes are more ancestral. We suggest the following model based on the information to date. The first eumetazoans had a complex of three genes (20, 21). Conventional thought considers these to represent head, trunk, and tail genes, although it seems likely that anterior-most development did not depend on the Hox genes. Duplication and divergence of the central gene ultimately gave rise to Dfd, Scr, and the Antp-class genes (Antp, Ubx, and abd-A) before the arthropod radiation (22). We propose that the ancestral trunk gene and its derivatives played two roles: suppression of genes resulting in anterior development and (probably in an evolutionarily labile fashion) determination of specialized trunk segment features. As noted, the beetle Antp, Scr, and Dfd orthologs all perform the former function in at least part of their expression domains [some evidence suggests that Drosophila homeotic genes play similar embryonic roles (3, 4, 23)]. On the other hand, it appears that the Tribolium Scr and Ubx orthologs no longer have this function in the thorax. That is, loss of function of the Antp ortholog is sufficient to transform the thoracic segments to antenna despite the normal expression of the Scr and Ubx orthologs in the anterior and posterior thorax, respectively (ref. 24 and unpublished observations). No data from flies or other arthropods speak to a possible ancestral role of lab in repressing anterior development in the intercalary segment; further speculation awaits the isolation of a Tclab mutation.
Acknowledgments
We thank Katherine Hummels and M. Susan Haas for technical assistance and Teresa Shippy for comments on the manuscript. This work was supported by grants from the National Science Foundation and the National Institutes of Health, as well as by the United States Department of Agriculture. M.De C. was supported by Human Frontier Science Program fellowship no. LT-568/93.
Abbreviations
- BXC
bithorax complex
- T3
metathorax
- PS
parasegment
- HOMC
homeotic complex
Footnotes
References
- 1.Lewis E. Nature (London) 1978;276:141–152. [Google Scholar]
- 2.Hayes P H, Sato T, Denell R E. Proc Natl Acad Sci USA. 1984;81:545–549. doi: 10.1073/pnas.81.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Struhl G. J Embryol Exp Morphol. 1983;76:297–331. [PubMed] [Google Scholar]
- 4.Sato T, Hayes P H, Denell R E. Dev Biol. 1985;111:171–192. [Google Scholar]
- 5.Rogers B T, Kaufman T C. Int Rev Cytol. 1997;174:1–84. doi: 10.1016/s0074-7696(08)62115-4. [DOI] [PubMed] [Google Scholar]
- 6.Jürgens G, Lehmann R, Schardin M, Nüsslein-Volhard C. Roux's Arch Dev Biol. 1986;195:359–377. doi: 10.1007/BF00402870. [DOI] [PubMed] [Google Scholar]
- 7.Stuart J J, Brown S J, Beeman R W, Denell R E. Nature (London) 1991;350:72–74. doi: 10.1038/350072a0. [DOI] [PubMed] [Google Scholar]
- 8.Brown S J, Holtzman S, Kaufman T, Denell R. Dev Genes Evol. 1999;209:389–398. doi: 10.1007/s004270050269. [DOI] [PubMed] [Google Scholar]
- 9.Beeman R W, Stuart J J, Haas M S, Friesen K S. J Hered. 1996;87:224–232. doi: 10.1093/oxfordjournals.jhered.a022989. [DOI] [PubMed] [Google Scholar]
- 10.Beeman R W, Stuart J J, Haas M S, Denell R E. Dev Biol. 1989;133:196–209. doi: 10.1016/0012-1606(89)90311-4. [DOI] [PubMed] [Google Scholar]
- 11.Brown S J, Mahaffey J P, Lorenzen M L, Denell R E, Mahaffey J W. Evol Dev. 1999;1:11–15. doi: 10.1046/j.1525-142x.1999.99013.x. [DOI] [PubMed] [Google Scholar]
- 12.Brown S J, Hilgenfeld R B, Denell R E. Proc Natl Acad Sci USA. 1994;91:12922–12926. doi: 10.1073/pnas.91.26.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González-Crespo S, Morata G. Development (Cambridge, UK) 1996;122:3921–3928. doi: 10.1242/dev.122.12.3921. [DOI] [PubMed] [Google Scholar]
- 14.Scholtz G, Mittmann B, Gerberding M. Int J Dev Biol. 1998;42:801–810. [PubMed] [Google Scholar]
- 15.Popadic A, Panganiban G, Rusch D, Shear W A, Kaufman T C. Dev Genes Evol. 1998;208:142–150. doi: 10.1007/s004270050165. [DOI] [PubMed] [Google Scholar]
- 16.McGinnis W, Jack T, Chadwick R, Regulski M, Bergson C, McGinnis N, Kuziora M A. Adv Genet. 1990;27:363–402. doi: 10.1016/s0065-2660(08)60030-9. [DOI] [PubMed] [Google Scholar]
- 17.O'Hara E, Cohen B, Cohen S M, McGinnis W. Development (Cambridge, UK) 1993;117:847–856. doi: 10.1242/dev.117.3.847. [DOI] [PubMed] [Google Scholar]
- 18.McGinnis N, Ragnhildstveit E, Veraksa A, McGinnis W. Development (Cambridge, UK) 1998;125:4553–4564. doi: 10.1242/dev.125.22.4553. [DOI] [PubMed] [Google Scholar]
- 19.Denell R E, Brown S J, Beeman R W. Semin Cell Dev Biol. 1996;7:527–538. [Google Scholar]
- 20.Finnerty J R. Curr Top Dev Biol. 1998;40:211–254. doi: 10.1016/s0070-2153(08)60368-3. [DOI] [PubMed] [Google Scholar]
- 21.Schierwater B, Kuhn K. Mol Phylogenet Evol. 1998;9:375–381. doi: 10.1006/mpev.1998.0489. [DOI] [PubMed] [Google Scholar]
- 22.Grenier J K, Garber T L, Warren R, Whitington P M, Carroll S. Curr Biol. 1997;7:547–553. doi: 10.1016/s0960-9822(06)00253-3. [DOI] [PubMed] [Google Scholar]
- 23.Röder L, Vola C, Kerridge S. Development (Cambridge, UK) 1992;115:1017–1033. doi: 10.1242/dev.115.4.1017. [DOI] [PubMed] [Google Scholar]
- 24.Beeman R W, Stuart J J, Brown S J, Denell R E. BioEssays. 1993;15:439–444. doi: 10.1002/bies.950150702. [DOI] [PubMed] [Google Scholar]