Abstract
There is an ongoing discussion on whether segmentation in different phyla has a common origin sharing a common genetic program. However, before comparing segmentation between phyla, it is necessary to identify the ancestral condition within each phylum. Even within the arthropods it is not clear which parts of the genetic network leading to segmentation are conserved in all groups. In this paper, we analyze the expression of three segmentation genes of the pair-rule class in the spider Cupiennius salei. Spiders are representatives of the Chelicerata, a monophyletic basic arthropod group. We find that in spider embryos, the orthologues for the Drosophila primary pair-rule genes hairy, even-skipped, and runt are expressed in stripes in the growth zone, where the segments are forming, suggesting a role for these genes in chelicerate segmentation. These data imply that the involvement of hairy, even-skipped, and runt in arthropod segmentation is an ancestral character for arthropods and is not restricted to a particular group of insects.
Metameric body plans are found in diverse metazoan phyla, including vertebrates, annelids, and arthropods. However, it is unclear whether the metamerization in different taxa is generated by a common mechanism (1). So far, the mechanisms underlying the segmentation process and the genes involved in this process have been studied most thoroughly in an insect, the fruit fly Drosophila (2, 3). In this species, segmentation genes were found to act in a hierarchical gene cascade, including gap genes, pair-rule genes, and segment-polarity genes. This gene cascade leads to the molecular subdivision of the embryo and, eventually, to the formation of the segments. The first sign of periodicity is the expression of the pair-rule genes; they are expressed in stripes in a double-segmental pattern, specifying alternate segments of the embryo. The overlapping stripes of the pair-rule genes then define the domains for the activity of the segment-polarity genes. These genes comprise the last step in the molecular hierarchy of the segmentation process (4). Within the arthropod phylum, the process of segmentation seems to be fairly conserved at the level of the segment-polarity genes (1). However, our knowledge of the genes acting farther upstream in the gene cascade is largely restricted to holometabolous insects, thus far.
Although apparent orthologues of the Drosophila segmentation genes have been found in many animal groups, including vertebrates, it is unclear to which extent they are involved in the generation of the repetitive patterns of segments and somites. In the beetle Tribolium castaneum, several pair-rule orthologues (hairy, even-skipped, fushi tarazu, and runt) have been identified and seem to be expressed in a double-segmental pattern (5–8). Furthermore, elimination of the Even-Skipped (EVE) protein in Tribolium results in a pair-rule phenocopy (9). In addition to this result, a systematic mutagenesis screen in Tribolium has uncovered mutants that display a clear pair-rule phenotype (10). This finding suggests a conserved function for at least some of the pair-rule genes in Tribolium. On the other hand, in the grasshopper Schistocerca, which is a representative of the more ancestral hemimetabolous insects, neither eve nor fushi tarazu (ftz) stripes form in the growth zone of the embryo (11, 12), suggesting that these genes do not act as pair-rule genes in this species.
To gain further insight into the evolution of the segmentation process and the role of segmentation genes in different animal phyla, it is necessary to identify the possible role of these genes within different taxa. For this reason, we analyzed the expression patterns of the pair-rule gene orthologues of hairy (h), even-skipped (eve), and runt (run) in the spider Cupiennius salei. These three genes are the primary pair-rule genes in Drosophila and are controlled directly by the gap genes. The basal phylogenetic position of chelicerates within the arthropod clade allows us to draw more general conclusions on the degree of conservation of this part of the segmentation-gene hierarchy. We find that Cs-h, Cs-eve, and Cs-run are expressed in stripes in the growth zone of the embryo, similar to what was found in Tribolium. This result suggests that the function of pair-rule genes in segmentation is ancestral, at least in arthropods.
Materials and Methods
Embryos.
We used embryos of the Central American wandering spider C. salei Keyserling (Chelicerata, Ctenidae, Aranida). Fertilized female spiders were obtained from a colony bred by Ernst-August Seyfarth in Frankfurt am Main, Germany. Embryos were collected as described before (13, 14).
Cloning of Cs-h, Cs-eve, and Cs-run.
Cs-h, Cs-eve, and Cs-run initially were found by reverse transcriptase (RT)-PCR on RNA prepared from germ-band embryos using degenerate primers directed against conserved positions in the basic helix–loop–helix region (hairy), the homeodomain (even-skipped), or the runt domain (runt). For Cs-h, we used the primers h-fw (AARCCNATHATGGARAARMGNMG) and h-bw-1 (YTGNARRTTYTGNARRTGYTTNAC) in an initial PCR, and h-fw and h-bw-2 (GTYWTYTCNARDATRTCNGCYTTYTC) in a nested PCR on a 1-μl aliquot of this initial PCR. For Cs-eve, we used the primers eve-fw (ACNGCNTTYACNMGNGARCA) and eve-bw-1 (CKYTGNCKYTTRTCYTTCAT) in an initial PCR, and eve-fw and eve-bw-2 (RTTYTGRAACCANACYTTDATNGT) in a nested PCR on a 1-μl aliquot of this initial PCR. For Cs-run, we used the primers run-fw-1 (RCNRYNATGAARAAYCARGTNGC) and run-bw (CKNGGYTCNCKNGGNCCRTC) in an initial PCR, and run-fw-2 (MRNTTYAAYGAYYTNMGNTTYGTNGG) and run-bw in a nested PCR on a 1-μl aliquot of this initial PCR. The obtained PCR fragments were cloned and sequenced.
A Cs-h cDNA clone was recovered from the embryonic cDNA library (13) by using the cloned PCR fragment as a probe to screen the library. Larger fragments for Cs-eve, covering the complete ORF, and for Cs-run, covering a part of the ORF, were obtained by rapid amplification of cDNA ends–PCR (Marathon cDNA amplification kit; CLONTECH).
Phylogenetic Analysis.
To produce the phylogenies, we first conducted a blast search (14) to identify the most closely related sequences in the data banks. From these, we selected a representative range of species. The sequences were aligned with clustal x (15) using the blosum matrix, a gap-opening penalty of 20, and a gap-extension penalty of 0.2. Phylogenetic analysis then was done with the program puzzle (16) as implemented in paup Version 4.0 (17).
In Situ Hybridization.
Whole mount in situ hybridizations were performed as described (13, 18), with the modifications for young stages as described in Damen and Tautz (19).
Results
Sequences of Cs-h, Cs-eve, and Cs-run.
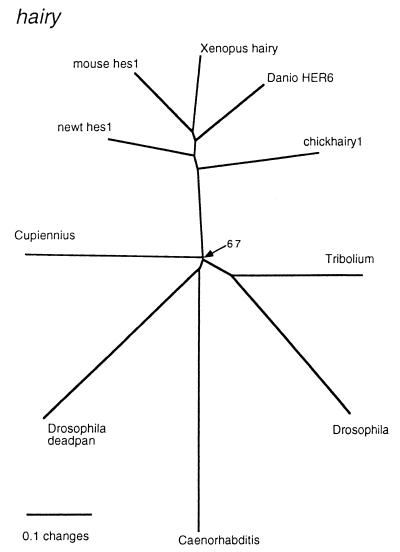
Initial RT-PCR amplification with the nested primers for hairy-like sequences yielded only one type of fragment with similarity to Drosophila hairy. This fragment was used to screen a cDNA library, and a 1.6-kb cDNA was recovered. The first ATG codon is found at nucleotide 141. However, because this is not preceded by an in-frame upstream stop codon, we cannot exclude the possibility that the amino-terminal part of the protein is missing. The phylogenetic analysis (Fig. 1) places the Cupiennius sequence at the base of the arthropods, but with the Caenorhabditis homologue and the Drosophila homologue deadpan inside this clade. However, this particular node is not strongly supported and is sensitive to the change of the alignment and reconstruction parameters.
Figure 1.
Phylogram of the sequences most related to Cupiennius hairy. All nodes had a puzzle support of >90%, except for the one that is specifically labeled. GenBank accession nos. for the sequences used: Cupiennius, AJ252154; Drosophila, S06956; Tribolium, S29712; Caenorhabditis, AF020555; Drosophila Deadpan, Q26263; mouse Hes1, NP032261; newt Hes1, BAA76633; Xenopus Hairy, AAA79185; Danio HER6, CAA65998; chickhairy1, O57337.
A remarkable feature of the Cs-H sequence is the change in the conserved carboxyl-terminal tetrapeptide WRPW found in the Hairy family of basic helix–loop–helix transcription factors, which include the Hairy, Deadpan, and Enhancer of split proteins. The WRPW tetrapeptide is changed to WRPF in Cs-H. The WRPW motif is required for interaction with the corepressor Groucho and for transcriptional repression (20, 21). We do not know whether this 1-aa change in the tetrapeptide affects a putative interaction of Cs-H with Groucho. Runt domain proteins contain a very similar carboxyl-terminal motif, WRPY, which also is required for Groucho-dependent repression in Drosophila (22).
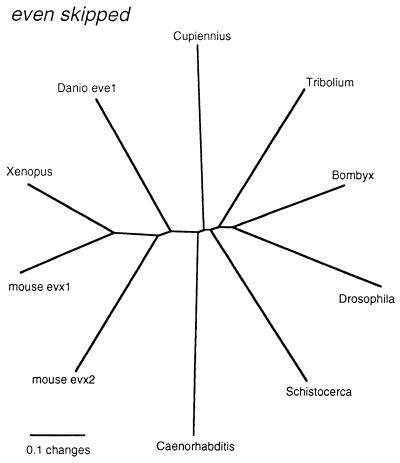
The initial RT-PCR amplification with the nested primers also yielded only one fragment for Cs-eve. The complete sequence for Cs-EVE was then obtained by 5′ and 3′ rapid amplification of cDNA ends–PCR experiments. Phylogenetic analysis places Cs-EVE within the other known EVE protein sequences, but at a basal position to the other arthropods (Fig. 2). All nodes are well resolved in this case and are in line with the accepted phylogenetic positions of each of the taxa.
Figure 2.
Phylogram of the sequences most related to Cupiennius even-skipped. All nodes had a puzzle support of >90%. GenBank accession nos. of the sequences used: Bombyx, D38486; Danio eve1, Q90265; Tribolium, P92067; Xenopus, P50476; mouse evx2, P49749; mouse evx1, P23683; Caenorhabditis, Q93899; Schistocerca, Z11845; Drosophila, P06602; Cupiennius, AJ252155.
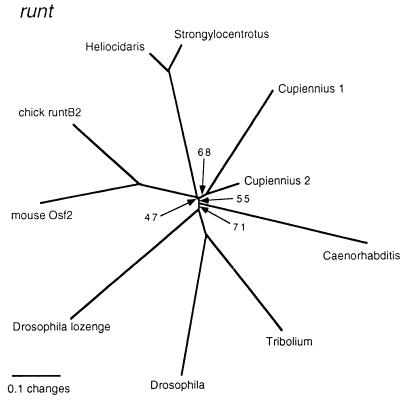
The initial RT-PCR amplification yielded two different types of PCR fragment for runt in the spider. For one of them (Cupiennius runt-1), we could obtain partial extensions of the 3′ and 5′ regions by rapid amplification of cDNA ends–PCR, which were used for the in situ hybridizations (see next paragraph). The phylogenetic analysis shows a poor resolution at the base of the tree (Fig. 3). There is moderate support for the two runt fragments from Cupiennius being related to each other. However, similar to the situation for hairy, we find the Caenorhabditis homologue and the Drosophila homologue lozenge inside the group that also contains the Drosophila and Tribolium runt genes. However, the exact order of these branches depends on the alignment and reconstruction parameters and therefore should be considered to be nonresolved.
Figure 3.
Phylogram of the sequences most related to the Cupiennius runt fragments. All nodes had a puzzle support of >90%, except for those that are specifically labeled. Note that only a small fragment was available for Cupiennius 2. GenBank accession nos. of the sequences used: Caenorhabditis, AAD54940; Drosophila, P22814; Drosophila lozenge, AAC47196; mouse Osf2, AAB65409; chick runtB2, CAA85297; Heliocidaris, AAC28443; Strongylocentrotus, AAB03565.
Expression of Cs-h, Cs-eve, and Cs-run.
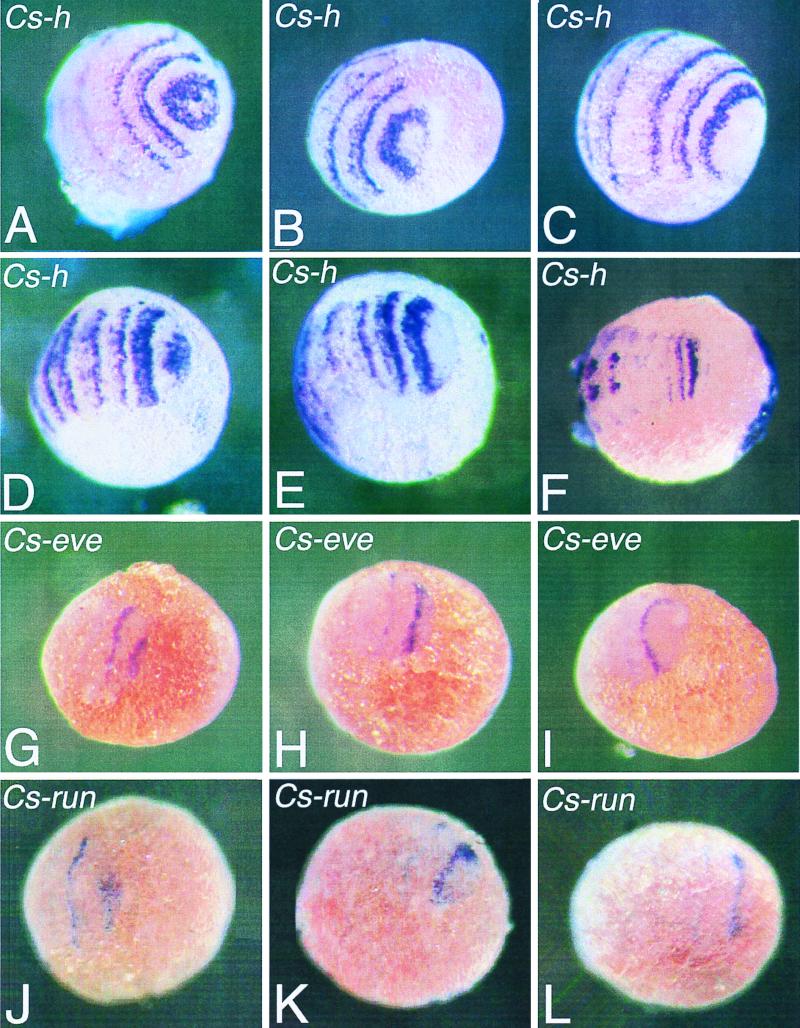
To see whether the spider pair-rule gene orthologues Cs-h, Cs-eve, and Cs-run play a role in the segmentation process of the spider, we analyzed their expression by whole mount in situ hybridization on young spider embryos. During spider embryogenesis, segments are sequentially added at the posterior end of the embryo, which resembles the formation of the abdominal segments in short-germ insect embryos. It appears that Cs-h, Cs-eve, and Cs-run are expressed in a dynamic way in stripes in the spider embryo. Fig. 4 A–C show expression of Cs-h in embryos before the segments of the prosoma (cephalothorax) become morphologically visible. Although the embryos are at comparable stages, they show distinct expression patterns that suggest a dynamic manner of stripe formation. Fig. 4A shows an embryo in which most of the posterior end is stained, whereas Fig. 4 B and C show a progressive clearing of this posterior end. A similar pattern can also be seen in later stages, alternate states of covering the most posterior end and clearance of it (Fig. 4 D and E). At more advanced stages of development, segments still form at the posterior end. This morphological segmentation still is preceded by hairy stripes (Fig. 4F).
Figure 4.
Embryos of the spider C. salei stained with the Cs-h (A–F), the Cs-eve (G–I), or the Cs-run (J–L) probe by whole mount in situ hybridization. Embryos in A–C are at a stage before the germ band is forming; embryos in D–E and G–L are slightly older, and the germ band is more compressed as compared with the embryos in A–C. The embryo in F is an even more advanced stage. The embryos in A–E, G–I, and J–L show the progression of the Cs-h, Cs-eve, and Cs-run expression, respectively. Initially, Cs-h, Cs-eve, and Cs-run are expressed in a posterior domain; these expression domains then move anteriorly and form stripes.
Cs-eve is also expressed in stripes that form from a posterior domain in a comparable way as Cs-h (Fig. 4 G–I), although the stripes are narrower and fade much faster. In the more differentiated segments, Cs-eve is then expressed in the developing central nervous system in each segment, a pattern that will be described in detail elsewhere.
Cs-run is expressed in a way that is very reminiscent of Cs-eve, in stripes in early embryos (Fig. 4 J–L). In addition, there is expression of this gene in the neuroectoderm of the more differentiated segments. Apart from this, Cs-run is also expressed in the head region of the developing spider embryo in a similar way as run is expressed in the head of the developing Drosophila embryo (23) and in the developing legs of the spider. These additional patterns will be described in detail elsewhere.
As soon as the broad Cs-h stripes become located more to the anterior, they fade at the anterior side and gain the appearance of a double stripe (Fig. 4 A–F). The anterior part of this double stripe is always weaker than the posterior. It is not clear yet whether this splitting of the broad Cs-h stripe is comparable to the splitting of eve stripes in Tribolium. In Tribolium, each primary eve stripe resolves in two segmental stripes (6, 24). For Cs-eve and Cs-run, we do not see such a splitting of the stripe.
The expression patterns in the spider demonstrate that morphological segmentation in the spider is preceded by expression of the segmentation gene orthologues Cs-h, Cs-eve, and Cs-run. Thus, Cs-h, Cs-eve, and Cs-run are likely to play a role in the segmentation process of the spider and might act upstream of the segment-polarity genes.
Discussion
Before one can understand the ancestral role of the segmentation genes during arthropod segmentation, it is necessary to compare the expression and function of these genes in different arthropod groups. The function of at least one gene is well conserved in arthropod segmentation, that of the segment-polarity gene engrailed (en). EN is expressed in the posterior part of the segments in insects, crustaceans, and chelicerates (13, 25–28), and it seems that it plays a similar role in the specification of segment borders. However, so far it is not clear whether the genes regulating en in the segmentation gene cascade are also conserved.
Our data show a likely involvement of three orthologues of insect pair-rule genes in spider segmentation. The expression of Cs-h, Cs-eve, and Cs-run precede morphological segmentation. This finding indicates that at least some of the upstream genes in the hierarchical segmentation gene cascade known from Drosophila do play a role in the segmentation of other arthropod clades. h, eve, and run belong to the primary pair-rule genes in Drosophila, which are directly controlled by the gap genes. Apart from the pair-rule genes, we also have recovered apparent orthologues of gap genes from spiders (ref. 29; our unpublished data). However, because the earliest stages of spider embryogenesis occur under conditions where the cells are only loosely aggregated, we have not yet been able to obtain expression patterns for these genes from the earliest stages. Thus, the involvement of gap genes and possibly of maternal genes in the segmentation of the spider embryo remains an open question. In addition to this, we are not yet able to say whether the pair-rule genes identified here act in a double-segmental periodicity. The Cs-h stripes resolve into segmental stripes at later stages, but this result could be a secondary expression aspect. Several pair-rule genes in Drosophila show both a double-segmental and a segmental periodicity. This phenomenon has been best documented for eve, where separate promoter elements drive the early and the late expression phases (30). It is interesting that evolutionary comparisons between insects show that this aspect of eve expression is rather variable. The mothmidge Clogmia (Diptera) shows only the pair-rule pattern, but not the segmental pattern (31). On the other hand, the parasitic wasp Copidosoma (Hymenoptera) shows only the segmental pattern (32). Both the double-segmental and segmental patterns are found in the honey bee (Hymenoptera) (33) and in Tribolium (Coleoptera) (6, 23). The more basic insect Schistocerca (Orthoptera) shows neither pattern, whereas a closely related orthopteran species, the cricket, as well as the earwig (Dermaptera), an even more basal insect, show a segmental pattern (P. Moore, R. Dawes, and N. Patel, unpublished data cited in ref. 1). These results suggest that not all components of the segmentation gene hierarchy need to be conserved in all species and that there may be a considerable flexibility for some genes. This flexibility could also explain the situation in Schistocerca, where neither eve nor ftz shows stripes in the growth zone (11, 12). However, both genes are expressed in a single domain in this region, providing the possibility that they still may be involved in some aspect of patterning regulation. It is also interesting that for ftz in Tribolium, loss of function is not associated with a pair-rule phenotype, although ftz is expressed in a pair-rule pattern in Tribolium (7, 34).
The fact that we find all three primary pair-rule genes known from Drosophila to be expressed in the growth zone of the spider suggests that the mechanism of patterning may be conserved, which would also imply that the pattern we see represents a pair-rule pattern, although we have no direct evidence for this assumption. This inference contrasts with the current knowledge about the formation of somites in vertebrates. It has been shown for the chick that somitogenesis is driven by an internal clock mechanism that provides each of the cells in the presomitic mesoderm with multiple waves of expression before a somite is specified (35–38). Intriguingly, one of the genes involved in this, chick hairy 1, is closely related to the arthropod hairy genes. On the other hand, it has been suggested for another gene from this family in zebrafish, her1, that it is expressed in a pair-rule pattern (39). Thus, it would seem possible that somitogenesis in vertebrates is driven by two mechanisms, a clock mechanism and a pair-rule mechanism. This possibility, in turn, could suggest that there is also a clock mechanism in arthropods that has not been detected yet, because most research has focused on understanding the pair-rule mechanism.
Acknowledgments
We thank Ernst-August Seyfarth (Frankfurt am Main, Germany) for providing us with mated spiders, Gabi Büttner for doing some of the in situ hybridizations, and Hilary Dove for critical reading of the manuscript. This work was supported by a Marie Curie Fellowship of the European Commission (Brussels) to W.D. and by a Deutsche Forschungsgemeinschaft grant (Ta99/15-1).
Abbreviation
- RT-PCR
reverse transcriptase–PCR
Footnotes
References
- 1.Davis G K, Patel N H. Trends Genet. 1999;15:M68–M72. [PubMed] [Google Scholar]
- 2.St. Johnston D, Nüsslein-Volhard C. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 3.Pankratz M, Jäckle H. In: Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 467–516. [Google Scholar]
- 4.Ingham P W. Nature (London) 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- 5.Sommer R J, Tautz D. Nature (London) 1993;361:448–450. doi: 10.1038/361448a0. [DOI] [PubMed] [Google Scholar]
- 6.Patel N H, Condron B G, Zinn K. Nature (London) 1994;367:429–434. doi: 10.1038/367429a0. [DOI] [PubMed] [Google Scholar]
- 7.Brown S J, Hilgenfeld R D, Denell R E. Proc Natl Acad Sci USA. 1994;91:12922–12926. doi: 10.1073/pnas.91.26.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown S J, Denell R E. Semin Cell Dev Biol. 1996;7:553–560. [Google Scholar]
- 9.Schröder R, Jay D G, Tautz D. Mech Dev. 1999;80:191–195. doi: 10.1016/s0925-4773(98)00211-1. [DOI] [PubMed] [Google Scholar]
- 10.Maderspacher F, Bucher G, Klingler M. Dev Genes Evol. 1998;208:558–568. doi: 10.1007/s004270050215. [DOI] [PubMed] [Google Scholar]
- 11.Patel N H, Ball E E, Goodman C S. Nature (London) 1992;357:339–342. doi: 10.1038/357339a0. [DOI] [PubMed] [Google Scholar]
- 12.Dawes R, Dawson I, Falciani F, Tear G, Akam M. Development (Cambridge, UK) 1994;120:1561–1572. doi: 10.1242/dev.120.6.1561. [DOI] [PubMed] [Google Scholar]
- 13.Damen W G M, Hausdorf M, Seyfarth E-A, Tautz D. Proc Natl Acad Sci USA. 1998;95:10665–10670. doi: 10.1073/pnas.95.18.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 17.Swofford D L. paup, Phylogenetic Analysis Using Parsimony. Washington, DC: Smithsonian Institution; 1998. , Version 4.064, prerelease version. [Google Scholar]
- 18.Damen W G M, Tautz D. Dev Genes Evol. 1998;208:586–590. doi: 10.1007/s004270050218. [DOI] [PubMed] [Google Scholar]
- 19.Damen W G M, Tautz D. J Exp Zool. 1999;285:85–91. doi: 10.1002/(sici)1097-010x(19990415)285:1<85::aid-jez10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 20.Jimenez G, Paroush Z, Ish-Horowicz D. Genes Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkhurst S M. Trends Genet. 1998;14:130–132. doi: 10.1016/s0168-9525(98)01407-3. [DOI] [PubMed] [Google Scholar]
- 22.Aronson B D, Fisher A L, Blechman K, Caudy M, Gergen J P. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kania M A, Bonner A S, Duffy J B, Gergen J P. Genes Dev. 1990;4:1701–1713. doi: 10.1101/gad.4.10.1701. [DOI] [PubMed] [Google Scholar]
- 24.Brown S J, Parrish J K, Beeman R W, Denell R E. Mech Dev. 1997;61:165–173. doi: 10.1016/s0925-4773(96)00642-9. [DOI] [PubMed] [Google Scholar]
- 25.Patel N H, Martin-Blanco E, Coleman K G, Poole S S, Ellis M C, Kornberg T, Goodman C S. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- 26.Scholtz G, Patel N H, Dohle W. Int J Dev Biol. 1994;38:471–478. [PubMed] [Google Scholar]
- 27.Patel N H. Development (Cambridge, U.K.) Suppl. 1994. , 201–207. [Google Scholar]
- 28.Telford M J, Thomas R H. Proc Natl Acad Sci USA. 1998;95:10671–10675. doi: 10.1073/pnas.95.18.10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sommer R J, Retzlaff M, Goerlich K, Sander K, Tautz D. Proc Natl Acad Sci USA. 1992;89:10782–10786. doi: 10.1073/pnas.89.22.10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goto T, MacDonald P, Maniatis T. Cell. 1989;57:413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- 31.Rohr K B, Tautz D, Sander K. Dev Genes Evol. 1999;209:145–154. doi: 10.1007/s004270050238. [DOI] [PubMed] [Google Scholar]
- 32.Grbic M, Nagy L M, Carroll S B, Strand M. Development (Cambridge, UK) 1996;122:795–804. doi: 10.1242/dev.122.3.795. [DOI] [PubMed] [Google Scholar]
- 33.Binner P, Sander K. Dev Genes Evol. 1997;7:447–454. doi: 10.1007/s004270050074. [DOI] [PubMed] [Google Scholar]
- 34.Stuart J J, Brown S J, Beeman R W, Denell R E. Nature (London) 1991;350:72–74. doi: 10.1038/350072a0. [DOI] [PubMed] [Google Scholar]
- 35.Palmeirim I, Henrique D, Ish-Horowicz D, Pourquié O. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- 36.Stern C D, Vasiliauskas D. BioEssays. 1998;20:528–531. doi: 10.1002/(SICI)1521-1878(199807)20:7<528::AID-BIES2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 37.McGrew M J, Dale J K, Fraboulet S, Pourquié O. Curr Biol. 1998;8:979–982. doi: 10.1016/s0960-9822(98)70401-4. [DOI] [PubMed] [Google Scholar]
- 38.Pourquié O. Curr Opin Genet Dev. 1999;9:559–565. doi: 10.1016/s0959-437x(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 39.Muller M, von Weizsacker E, Campos-Ortega J A. Development (Cambridge, UK) 1996;122:2071–2078. doi: 10.1242/dev.122.7.2071. [DOI] [PubMed] [Google Scholar]