Abstract
The goal of this study was to evaluate binding of four targets of biodefense interest to immobilized antimicrobial peptides (AMPs) in biosensor assays. Polymyxins B and E, melittin, cecropins A, B, and P, parasin, bactenecin and magainin-1, as well as control antibodies, were used as capture molecules for detection of Cy3-labeled Venezuelan equine encephalitis virus (VEE), vaccinia virus, C. burnetti and B. melitensis. Although VEE, vaccinia virus and C. burnetti did not show any binding activity to their corresponding capture antibodies, B. melitensis bound to immobilized anti-Brucella monoclonal antibodies. The majority of the immobilized AMPs included in this study bound labeled VEE, vaccinia virus and C. burnetti in a concentration-dependent manner, and B. melitensis bound to polymyxin B, polymyxin E, and bactenecin. No binding was observed on immobilized magainin-1. In contrast to all bacterial targets tested to date, VEE and vaccinia virus demonstrated similar patterns of binding to all peptides. While the direct assay is generally replaced by a sandwich assay for analysis of real-world samples, direct binding experiments are commonly used to characterize specificity and sensitivity of binding molecules. In this case, they clearly demonstrate the capability of AMPs as recognition molecules for four biothreat agents.
Keywords: antimicrobial peptides, Coxiella burnetti, Brucella melitensis, vaccinia virus, Venezuelan equine encephalitis virus, screening
INTRODUCTION
Biological warfare has evolved from use of blood, manure, and decomposing tissues on arrowheads to modern day, state-run programs for development of large-scale deployable biological weapons [1]. As early as 1970, the World Health Organization has publicly recognized the high human cost inherent in biological agent attacks [2]. Economic costs associated with an intentional or accidental biological agent release are also significant; the Centers for Disease Control has estimated the economic impact of a bioterrorist attack, ranging from $477 million to $26 billion per 100,000 persons exposed, depending on the agent used [3].
Still considered the “gold standard” for detection/diagnosis of exposure and infection, culture of live bacterial or viral threat agents is time-consuming, requires significant technical skill and, depending on the agent, biosafety level 3 or 4, thereby necessitating transport to an appropriate laboratory facility. Although rapid detection systems such as lateral flow devices are still useful for on-site detection, enzyme-linked immunosorbent assays (ELISAs) and real-time PCR are the most helpful tests in a clinical setting. The key advantages of ELISA are its overall sensitivity and the lack of requirement for significant sample preparation. PCR, on the other hand, is generally more sensitive and specific and, once nucleic acids are extracted, rapid; a number of excellent, high sensitivity PCR-based assays have been described [4-11]. However, both antibody- and nucleic acid-based detection methods require prior development of target-specific reagents whose efficacy is highly dependent on assumed genetic sequences and/or antigenic motifs. Changes as simple as modification of growth conditions or as complex as genetic manipulations may confound results. In an effort to circumvent some of these limitations, we have explored alternative recognition molecules for use in detection assays as a complementary technique to standard immunoassays. In particular, these alternative recognition species may be highly beneficial in cases where development of sensitive immunoassays has proven challenging.
This paper describes assays for four inactivated targets of biodefense interest using non-antibody-based recognition. Target recognition in these assays was accomplished using immobilized antimicrobial peptides (AMPs). AMPs comprise part of the innate immune system of many organisms and help protect the host species from microbial invasion/infection. Current dogma dictates that the antimicrobial activity of these peptides is mediated through interaction of AMPs with target cell membranes and subsequent membrane disruption, although other mechanisms have been postulated [12-14]. The current study expands on previous work developing AMP-based detection assays for Escherichia coli and Salmonella typhimurium [15,16]. As a first step in development of a multiplexed system for biothreat detection, direct assays for two intracellular pathogens (Coxiella burnetti, Brucella melitensis) and two enveloped viruses (Venezuelan equine encephalitis [VEE], vaccinia virus) were demonstrated using fluorescently labeled targets.
EXPERIMENTAL
Reagents
InactivatedC. burnetti and B. melitensis cells, VEE, vaccinia virus, and their corresponding antibodies were obtained from the US Department of Defense Critical Reagent Program; cells and viruses were certified non-infectious prior to shipment. Antimicrobial peptides cecropin A and B and magainin-1 were received from Anaspec (San Jose, CA), polymyxin B and E were from Sigma-Aldrich (St. Louis, MO), melittin, cecropin P, bactenicin and parasin were from American Peptides (Sunnyvale, CA). Sequences of the peptides are present in Table 1. All peptides were used as received without additional purification procedures.
Table 1.
Amino acid sequences of AMPs used in this study.
| Polymyxin B | |
| Polymyxin E | |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ-CONH2 |
| Cecropin A | KWKLFKKIEKVGQNIRDGIIKAGPAVAVVGQATQIAK-CONH2 |
| Cecropin B | KWKVFKKIEKMGRNIRNGIVKAGPAIAVLGEAKAL |
| Cecropin P | SWLSKTAKKLENSAKKRISEG IAIAIQGGPR |
| Bactenecin | |
| Magainin-1 | GIGKFLHSAGKFGKAFVGEIMKS |
| Parasin | KGRGKQGGKVRAKAKTRSS |
Preparation of fluorescent cells
Cells or viral particles were diluted to approximately 108 cells/ml or 109 pfu/ml in 50 mM sodium borate, pH 8.5. Each cell or viral suspension was then incubated with one packet of Cy3 bisfunctional N-hydroxysuccinimidyl ester (Amersham, Arlington Heights, IL, sufficient to label 1 mg protein), previously dissolved in 25μL anhydrous dimethyl sulfoxide. After 30 min incubation at room temperature, the labeled cells/viruses were loaded into dialysis tubing (1000 MWCO) and dialyzed overnight at 4°C against phosphate buffered saline, pH 7.4 (PBS) with 3 changes of buffer [15]. The labeled species were then stored in the dark at 4°C until use.
Substrate preparation and immobilization of peptides and antibodies
The detailed description of slide preparation for immobilization was described elsewhere [17]. Briefly, standard microscope slides (Daigger, Vernon Hills, IL) were cleaned with 10% KOH (w/v) in methanol for 1 hour, rinsed extensively with deionized water, dried under nitrogen, and treated with a 2% solution of 3-mercaptopropyl trimethoxysilane in toluene for 1 hour. Then the slides were extensively rinsed with toluene, dried under nitrogen, and incubated for 30 minutes in 1 mM N-[γ-maleimidobutyryloxy]-succinimide ester (Pierce, Rockford, IL) in absolute ethanol. After rinsing briefly in water and drying under nitrogen, the slides were placed in contact with poly(dimethyl)siloxane (PDMS) patterning templates molded to possess 15-channels oriented across the short (25 mm) axis of the slide. Capture solutions containing AMPs or antibodies diluted in PBS were injected into the PDMS channels and incubated overnight at 4°C. Concentrations of the recognition molecules in the capture solutions were as follows: Polymyxins B and E, 10 mg/ml; cecropins A, B and P, magainin-1, and parasin, 1 mg/ml; melittin and bactenecin, 250 μg/ml; control and target-specific antibodies, 10 μg/ml. After overnight incubation with the capture solutions, the channels were emptied and rinsed with PBS. The slides were then blocked for 30 min in 10 mg/ml gelatin in PBS, dried, and stored at 4°C for up to 2 weeks.
Assay Protocol
Patterned slides were placed in contact with PDMS assay templates molded to contain 12 channels oriented orthogonal to the channels in the patterning templates. Prior to the assay, each channel was first rinsed with 1 ml of PBS containing 1 mg/ml bovine serum albumin and 0.05% Tween-20 (PBSTB) at 0.8 ml/min using a multichannel peristaltic pump. Samples of Cy3-labeled species (0.1 ml, diluted in PBSTB) were then injected into appropriate channels and allowed to incubate for 1 hr at room temperature in the dark. Each channel was then washed with 1 ml of PBSTB, under flow (0.3 ml/min), using a multichannel peristaltic pump. After removing the PDMS templates, the slides were washed with deionized water, dried under nitrogen, and imaged with a Packard ScanArray Lite confocal microarray scanner (Packard Biochip Technologies, Billerica, MA).
Fluorescence imaging and data analysis
Fluorescence intensities were extracted from the images using QuantArray microarray analysis software Program (Packard). Concentration-dependence curves were fitted to a 3-parameter exponential rise to maximum function using Sigma Plot's automated curve fit (Sigma Plot software, Version 8.0, Chicago, IL). Signals for binding to various AMPs (mean ± standard deviation, n ≥ 3) were normalized with respect to positive controls (chicken antibody) to account for variability between the slides. Detection limits (LODs) were calculated as lowest tested concentrations giving signals at least 3 standard deviations above the mean of negative control values (n ≥ 3).
RESULTS
The original goal of this study was to develop binding assays of VEE, vaccinia virus, C. burnetti, and B. melitensis on AMPs using sandwich and direct assay formats (Figure 1). No significant binding of any of the four targets to immobilized AMPs or their corresponding antibodies was observed when sandwich-format assays were used (data not shown). These results indicated that either the fluorescent tracer antibodies or the capture species (antibodies, AMPs) lacked sufficient affinity for detection at the concentrations of targets tested. However, direct binding of fluorescently labeled targets to immobilized AMPs, but not to the corresponding “capture” antibodies, was observed, indicating that the problem in the sandwich assays was antibody-related. To eliminate the role of antibodies in AMP-based assays for VEE, vaccinia virus, C. burnetti, and B. melitensis, the direct assay approach was used in all further studies.
Figure 1.
Schematic of direct (left) and AMP-sandwich (right) assay formats. In both formats, AMPs are covalently immobilized to the planar substrate as “capture” reagents.
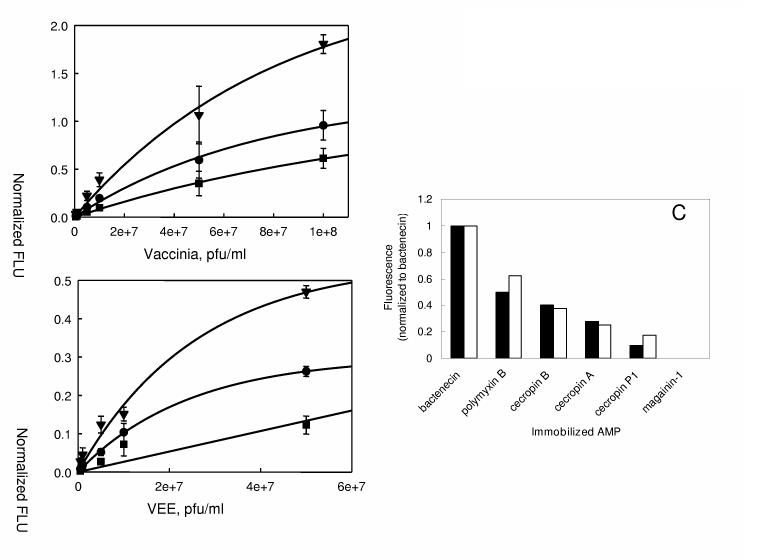
Figure 2 shows the images (Panel A) and concentration-dependence curves (Panel B) of Cy3-labeled VEE and vaccinia virus binding to several AMPs. Although the viruses did not bind significantly to their own specific antibodies, there was a small, but statistically significant degree of non-specific binding of both VEE and vaccinia virus to the opposite antibodies at the highest concentration tested (5 × 107 pfu/ml). In general, AMP-based assays for vaccinia virus showed higher sensitivity than those for VEE, although this effect was AMP-dependent. Detection limits are presented in Table 2. No binding of either virus was observed to magainin-1 at the concentration range used in this study. Figure 2B compares binding of VEE and vaccinia virus to bactenecin (dashed line) and polymyxin B (solid line) as examples of two AMPs used in this study.
Figure 2.
A. Confocal scanner images of Cy3-labeled viruses binding to immobilized AMPs on the sensing substrate. Control antibodies for VEE and vaccinia virus are shown abbreviated as anti-VEE and anti-Vac, respectively. PBS served as a negative (−) control and anti-chicken IgY as a positive (+) control. Incorporation of anti-chicken IgY allowed comparison of slides between independent experiments. B. Concentration-dependence curves for Cy3-labeled VEE (○) and vaccinia virus (●) binding to immobilized polymyxin B (solid line) and bactenecin (dashed line). Error bars indicate standard deviation (n = 3 slides). Fluorescent intensities were normalized with respect to chicken positive controls.
Table 2.
Detection limits for Cy3-labeled species on immobilized AMPs and corresponding antibodies.
| Recognition molecule | Detection limits | |||
|---|---|---|---|---|
| VEE pfu/ml | Vaccinia virus pfu/ml | C. burnetti cells/ml | B. melitensis cells/ml | |
| Polymyxin B | 5 × 106 | 5 × 105 | 5 × 105 | 5 × 106 |
| Polymyxin E | 5 × 106 | 5 × 105 | 5 × 105 | 5 × 106 |
| Melittin | 5 × 106 | 5 × 105 | 5 × 105 | > 5 × 106 |
| Cecropin A | 5 × 106 | 1 × 106 | 1 × 106 | > 5 × 106 |
| Cecropin B | 1 × 106 | 5 × 105 | 5 × 105 | > 5 × 106 |
| Cecropin P | 1 × 107 | 5 × 106 | 5 × 105 | > 5 × 106 |
| Bactenecin | < 5 × 105(a) | < 5 × 105 (a) | 5 × 105 | 5 × 104 (b) |
| Parasin | 1 × 107 | 5 × 106 | 5 × 105 | > 5 × 106 |
| Magainin-1 | > 1 × 108 | 1 × 107 | > 1 × 107 | > 5 × 106 |
| Corresponding antibody | > 1 × 108 | > 1 × 108 | > 1 × 107 | 5 × 105(c) |
lower concentrations were not tested in this study
some inconsistency in B. melitensis binding to bactenicin was noted from slide to slide.
binding was observed only when monoclonal antibody was used as a capture molecule.
Figure 3 shows concentration-dependence plots for the binding of vaccinia virus and VEE to the cecropin group of AMPs - cecropins A, B and P. In all cases, the immobilized AMPs gave higher signals and lower detection limits for binding to vaccinia virus compared to VEE (see also Table 2). Cecropin P demonstrated lowest binding ability among the three cecropins to capture labeled viruses and binding did not reach saturation for range of concentrations of both VEE and vaccinia virus used here. Affinities of the two viral species to immobilized parasin and melittin were similar to those of cecropin P and cecropin B, respectively. Overall, although different sensitivities were observed in vaccinia virus and VEE assays, both viruses showed virtually superimposable patterns of binding to the immobilized AMPs (Figure 3C).
Figure 3.
Concentration-dependence curves for Cy3-labeled vaccinia virus (A) and VEE (B) binding to cecropin A (●), cecropin B (▼) and cecropin P (■). Error bars indicate standard deviation (n=3 slides). Fluorescent intensities were normalized with respect to chicken positive control. Panel C: Patterns of binding of vaccinia virus (closed bars) and VEE (open bars) normalized to bactenecin results.
Figure 4 shows the image (right panel) and concentration-dependence plots (Panels A and B) for Cy3-labeled C. burnetti binding to immobilized AMPs. Consistent with VEE and vaccinia virus, no binding was observed to the immobilized Coxiella-specific antibody. However, there was some non-specific binding to anti-B. melitensis antibody and negative controls (PBS lane) at high concentrations (5 × 106 cells/ml and above, highest concentrations not shown on the image). Figure 4 shows concentration-response curves for C. burnetti binding to bactenecin and polymyxin B (Panel A) and the three cecropins (Panel B), analogous to the curves shown in Figures 2 and 3 for VEE and vaccinia virus. In contrast to the viral targets, polymyxin B demonstrates higher affinity than bactenecin for binding to C. burnetti. Polymyxin E showed similar affinity for C. burnetti as polymyxin B, with melittin and cecropin B showing somewhat lower affinities. Binding curves for C. burnetti to cecropin A, cecropin P1, and parasin (data not shown) were not significantly different from each other (Panel B).
Figure 4.
Confocal scanner image of Cy3-labeled C. burnetti binding to immobilized AMPs (right). Control antibody for C. burnetti is shown abbreviated as anti-Cox. PBS served as a negative (−) control and anti-chicken IgY as a positive (+) control. Incorporation of anti-chicken IgY allowed comparison of slides between independent experiments. A. Concentration-dependence curves for Cy3-labeled C. burnetti binding to polymyxin B (○) and bactenecin (●). B. Concentration-dependence curves for Cy3-C. burneti binding to cecropin A (●), cecropin B (▼) and cecropin P (■). Error bars indicate standard deviation (n = 4 slides). Fluorescent intensities were normalized with respect to chicken positive control.
B. melitensis demonstrated a low level of binding to AMPs, as well as to its own specific antibody and bound only to polymyxins B and E and bactenecin (Table 2). Although labeled Brucella cells bound to immobilized bactenecin, quantitative results varied from slide to slide. With some degree of variability, B. melitensis was the only species that showed binding to corresponding antibody that was, in some cases, significantly above background (P < 0.05); this result was observed only using monoclonal antibodies (data not shown).
DISCUSSION
Three of the four targets investigated in the present study (C. burnetti, B. melitensis, and VEE) have been cited as possible weapons for use against humans by numerous international committees [2, 18, 19]; the fourth target, vaccinia virus, is a simulant for variola, the etiologic agent of smallpox.
As a number of AMPs have been demonstrated to have antiviral properties, two enveloped viruses of biodefense interest were included in this study. Potentially life-threatening itself in immunocompromised individuals, vaccinia virus was used in this study as a surrogate for variola virus. Variola is considered a CDC Category A pathogen due to the potential susceptibility of many populations, its high mortality rate (up to 30%), and the lack of specific treatment. It is highly transmissible through contact and aerosols and was researched for weaponization. VEE is also stable and highly infectious by aerosol with great potential for causing epidemic human disease [20] with prolonged incapacitation and convalescence.
Detection limits for vaccinia virus (5 × 105 pfu/ml) on the AMP array were significantly higher than those obtained using a cell-based system [21] and an antibody-based array platform [22], as well as PCR-based tests [9, 11]. However, detection limits determined here were in the same range as another biosensor-based assay that does not utilize concentration or amplification steps [23]. Although assays were less sensitive for VEE, detection limits for VEE in the AMP-based assays were similar to or better than those of competing cell- or antibody-based technologies [21, 22, 24, 25].
Several groups have previously described the antiviral effects of various AMPs and their derivatives on vaccinia virus [26-28]. Most relevant to the present study is a cecropin A-magainin-2 hybrid peptide (CA(1-8)-MA(1-12)) which was shown to exhibit antiviral effects in other studies [26]. The three cecropins (with identical sequences in the first 8 amino acids) exhibited varying affinities for vaccinia virus in the present study, thereby implicating the positions of charged residues and lengths of hydrophobic segments within the C-terminal sequences for these differences. Magainin-1, whose N-terminal sequence differs from that used in CA(1-8)-MA(1-12) by a single residue, was not bound by either virus at concentrations up to 108 pfu/ml, potentially implicating its own C-terminal sequence for differences in activities between the two studies. Inclusion of CA(1-8)-MA(1-12), magainin-2, and derivatives thereof in future studies may help elucidate the mechanistic differences in binding and antiviral activity.
Brucella spp. and C. burnetti are the etiologic agents for the zoonotic diseases brucellosis and Q fever, respectively. Both B. melitensis and C. burnetti are intracellular pathogens and, though not generally considered high lethality species, both can cause long-term debilitating illnesses. Environmentally stable in dried form, both species are extremely infectious by the airborne route; inhalation of as few as 1-100 cells of either species can produce disease [29-33]. Although bacteremia may be present in early stages of infection with both agents, diagnosis is typically made through serology, in large part due to the low numbers of circulating cells, as well as the relative lack of sensitivity of immunoassays for direct detection of the causative organisms.
B. melitensis exhibited a low level of binding to both its immobilized capture antibody and immobilized bactenecin and polymyxins in direct assays, but no significant binding to the antibody was observed in sandwich assays. The limit of detection on bactenecin (5 × 104 cfu/ml) was 10-fold higher than observed in two other rapid biosensor systems employing antibodies for detection [34, 35], but 20-fold higher than an additional array-based method [22]. Not surprisingly, B. melitensis did not demonstrate any significant binding to immobilized cecropins, magainin-1, or melittin; the Brucella strain used in this study produces “smooth” lipopolysaccharide (LPS) and several researchers have documented that smooth strains of Brucella are significantly more resistant than rough strains to inactivation/killing by these specific AMPs [36-38]. Interestingly, B. melitensis bound to bactenecin to a higher extent than to polymyxin B in the present study, whereas Martinez de Tejada and coworkers [37] observed the opposite effect on viability; however, it has been demonstrated that AMP-membrane binding does not always result in cell killing [39].
During growth, C. burnetti assumes two different phases where the O-antigen chain and some components of the outer core of LPS vary. The Phase I form used here is highly virulent and corresponds to the “smooth” LPS variants of Brucella. However, in contrast to results with the B. melitensis smooth strain, C. burnetti showed significant affinity for a number of the immobilized AMPs. Although not as sensitive as PCR for detection of C. burnetti [10, 40], the detection limits in the AMP-based assays were within an order of magnitude as those from an ELISA previously [41]. Polymyxin B appeared to be a more potent capture reagent at higher cell numbers and has previously been demonstrated to interact with C. burnetti Phase I LPS [42, 43]. However, bactenecin bound more cells at lower cell concentrations. Interestingly, an unrelated cationic antimicrobial peptide, CAP37(20-44) [sequence 20-NGQRHFC*GGALIHARFVMTAASC*FQ-44, disulfide bond between C26 and C42] was shown to enhance the C. burnetti infectivity in mouse fibroblast cells [44]; this peptide possess several motifs in common with bactenecin: an intramolecular disulfide bond, a mixture of hydrophobic and cationic amino acids within the looped structure, several arginine residues, and the presence highly polar residues at both termini. It will be interesting in future experiments to determine if other peptides with these and similar motifs show strong affinity for C. burnetti cells and how binding is affected by phase-related changes in LPS structure.
The research described here is an initial step in the development and implementation of a multi-analyte detection assay for targets of biodefense interest. A key characteristic of AMPs, expected to be advantageous in the final embodiment of these assays, is their overlapping specificities and the ability to bind multiple targets with differing affinities. An array-based system using multiple AMPs for recognition would potentially be able to detect a larger number of targets than a corresponding number of antibodies, with identification of the target based on its pattern of binding. As observed with all other bacterial targets tested to date [15, 16, unpublished], both C. burnetti and B. melitensis showed clear differences in binding to the immobilized AMPs. However, in spite of differences in the magnitudes of binding, the patterns of binding of VEE and vaccinia virus to multiple immobilized AMPs were very similar and could not be distinguished. This could potentially be a unique characteristic for viruses, but may depend on the cells used for growth; a thorough study of the growth conditions of these and other viral strains should help determine whether AMP-based assays may find use for identification of viral targets.
Finally, although assays requiring labeled targets are not the optimal format for rapid detection in real-world scenarios, this study has shown that immobilized AMPs can be used to detect viral, bacterial, and rickettsial targets. As we have previously encountered difficulties in obtaining high affinity antibodies directed against these targets, the use of AMPs as both capture molecules and labeled “tracers” is presently being explored. Use of a universal or pan-specific tracer species, or a non-specific labeling method [45] could potentially allow detection of these and many other heretofore challenging species with high sensitivity, with the possibility of identification based on pattern recognition.
ACKNOWLEDGEMENTS
N.V.K. was recipient of National Research Council research associate fellowship. The authors wish to thank the National Institutes of Health Grant EB000680 and the US Department of Defense for their financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lesho E, Dorsey D, Bunner D. Feces, dead horses, and fleas: Evolution of the hostile use of biological agents. West. J. Med. 1998;168:512–516. [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Group of consultants . Health aspects of chemical and biological weapons. World Health Organization; Geneva, Switzerland: 1970. [Google Scholar]
- 3.Kaufmann AF, Meltzer MI, Schmid GP. The economic impact of a bioterrorist attack: are prevention and past attack programs justifiable? Emerg. Infect. Dis. 1997;3:83–94. doi: 10.3201/eid0302.970201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fenollar F, Fournier PE, Raoult D. Molecular detection of Coxiella burnetti in sera of patients with Q-fever endocarditis or vascular infection. J. Clin. Microbiol. 2004;42:4919–1924. doi: 10.1128/JCM.42.11.4919-4924.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navarro E, Escribano J, Fernandez J, Solera J. Comparison of three different PCR methods for detection of Brucella spp. in human blood samples. FEMS Immunol. Med. Microbiol. 2002;34:147–151. doi: 10.1111/j.1574-695X.2002.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 6.Newby DT, Hadfield TL, Roberto FF. Real-time PCR detection of Brucella abortus: A comparative study of SYBR Green I, 5′-exonuclease, and hybridization probe assays. Appl. Environ. Microbiol. 2003;69:4753–4759. doi: 10.1128/AEM.69.8.4753-4759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen DR, Hartman LJ, Loveless BM, Frye MS, Shipley MA, Bridge DL, Richards MJ, Kaplan RS, Garrison J, Baldwin CD, Kulesh DA, Norwood DA. Detection of biological threat agents by real-time PCR: comparison of assay performance on the R.A.P.I.D., the LightCycler, and the SmartCycler platforms. Clin. Chem. 2006;52:141–145. doi: 10.1373/clinchem.2005.052522. [DOI] [PubMed] [Google Scholar]
- 8.Linssen B, Kinney RM, Aguilar P, Russell KL, Watts DM, Kaaden OR, Pfeffer M. Development of reverse transcription-PCR assays specific for detection of equine encephalitis viruses. J. Clin. Microbiol. 2000;38:1527–1535. doi: 10.1128/jcm.38.4.1527-1535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espy MJ, Cockerill FR, III, Meyer RF, Bowen MD, Poland GA, Hadfield TL, Smith TF. Detection of smallpox virus DNA by LightCycler PCR. J. Clin. Microbiol. 2002;40:1985–1988. doi: 10.1128/JCM.40.6.1985-1988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klee SR, Tyczka J, Ellerbrok H, Franz T, Linke S, Baljer G, Appel B. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetti. BMC Microbiol. 2006;6:2. doi: 10.1186/1471-2180-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nitsche A, Steger B, Ellerbrok H, Pauli G. Detection of vaccinia virus DNA on the LightCycler by fluorescence melting curve analysis. J. Virol. Methods. 2005;126:187–195. doi: 10.1016/j.jviromet.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 13.Haukland HH, Ulvatne H, Sandvik K, Vorland LH LH. The antimicrobial peptides lactoferricin B and magainin 2 cross over the bacterial cytoplasmic membrane and reside in the cytoplasm. FEBS Lett. 2001;508:388–393. doi: 10.1016/s0014-5793(01)03100-3. [DOI] [PubMed] [Google Scholar]
- 14.Hsu CH, Chen C, Jou ML, Lee AY, Lin YC, Yu YP, Huang WT, Wu SH. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucl. Ac. Res. 2005;33:4053–4064. doi: 10.1093/nar/gki725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulagina NV, Lassman ME, Ligler FS, Taitt CR. Use of an antimicrobial peptide for detection of bacteria in biosensor assays. Anal. Chem. 2005;77:6504–6508. doi: 10.1021/ac050639r. [DOI] [PubMed] [Google Scholar]
- 16.Kulagina NV, Shaffer KM, Anderson GP, Ligler FS, Taitt CR. Antimicrobial peptide-based array for Escherichia coli and Salmonella screening. Anal. Chim. Acta. 2006;575:9–15. doi: 10.1016/j.aca.2006.05.082. [DOI] [PubMed] [Google Scholar]
- 17.Ngundi MM, Taitt CR. An array biosensor for detection of bacterial and toxic contaminants of foods. Meth. Mol. Biol. 2006;345:53–68. doi: 10.1385/1-59745-143-6:53. [DOI] [PubMed] [Google Scholar]
- 18. United Nations Security Council document S/23500, 31 January 1992.
- 19. Australia Group document AG/Dec92/BW/Chair/30 dated June 1992.
- 20.Smith JF, Davis K, Har MK, Ludwig GV, McClain DJ, Parker MD, Pratt WD. In: Medical Aspects of Chemical and Biological Warfare. Sidell FR, Takajufi ET, Franz DR, editors. Office of the Surgeon General; Washington, DC: 1999. pp. 561–589. Chapter 28. [Google Scholar]
- 21.Rider TH, Petrovic MS, Nargi RE, Harper JD, Schwoebel ED, Mathews RH, Blanchard DJ, Bortolin LT, Young AM, Chen J, Hollis MA. A cell-based sensor for rapid identification of pathogens. Science. 2003;301:213–215. doi: 10.1126/science.1084920. [DOI] [PubMed] [Google Scholar]
- 22.Huelseweh B, Ralf Ehricht R, Marschall H-J. A simple and rapid protein array based method for the simultaneous detection of biowarfare agents. PROTEOMICS. 2006;6:2972–2981. doi: 10.1002/pmic.200500721. [DOI] [PubMed] [Google Scholar]
- 23.Donaldson KA, Kramer MF, Lim DV. A rapid detection method for vaccinia virus, the surrogate for smallpox virus. Biosens. Bioelectron. 2004;20:322–327. doi: 10.1016/j.bios.2004.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu W-G, Thompson HG, Alvi AZ, Nagata LP, Suresh MR, Fulton RE. Development of immunofiltration assay by light addressable potentiometric sensor with genetically biotinylated recombinant antibody for rapid identification of Venezuelan equine encephalitis virus. J. Immunol. Methods. 2004;289:27–35. doi: 10.1016/j.jim.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Smith DR, Rossi CA, Kijek TM, Henchal EA, Ludwig GV. Comparison of dissociation-enhanced lanthanide fluorescent immunoassays to enzyme-linked immunosorbent assays for detection of staphylococcal enterotoxin B, Yersinia pestis-specific F1 antigen, and Venezuelan equine encephalitis virus. Clin. Diagn. Lab. Immunol. 2001;8:1070–1075. doi: 10.1128/CDLI.8.6.1070-1075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee GD, Park Y, Lin I, Hahm K-S, Lee H-Y, Moon Y-H, Woo E-R. Structure-antiviral activity relationships of cecropin A-magainin 2 hybrid peptide and its analogues. J. Peptide Sci. 2004;10:298–303. doi: 10.1002/psc.504. [DOI] [PubMed] [Google Scholar]
- 27.Howell MD, Jones JF, Kisich KO, Streib JE, Gallo RL, Leung DYM. Selective killing of vaccinia virus by LL-37: Implications for eczema vaccinatum. J. Immunol. 2004;172:1763–1767. doi: 10.4049/jimmunol.172.3.1763. [DOI] [PubMed] [Google Scholar]
- 28.Gordon YJ, Huang LC, Romonoswki EG, Yates KA, Proski RJ, McDermott AM. Human cathelicidin (LL-37), a multifunctional peptide, is expressed by ocular surface epithelia and has potent antibacterial and antiviral activity. Curr. Eye Res. 2005;30:385–394. doi: 10.1080/02713680590934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrne WR. In: Medical Aspects of Chemical and Biological Warfare. Sidell FR, Takajufi ET, Franz DR, editors. Office of the Surgeon General; Washington, DC: 1999. pp. 523–537. Chapter 26. [Google Scholar]
- 30.Hoover DL, Friedlander AM. In: Medical Aspects of Chemical and Biological Warfare. Sidell FR, Takajufi ET, Franz DR, editors. Office of the Surgeon General; Washington, DC: 1999. pp. 513–521. Chapter 28. [Google Scholar]
- 31.Cutler SJ, Whatmore AM, Commander NJ. Brucellosis – new aspects of an old disease. J. Appl. Microbiol. 2005;98:1270–1281. doi: 10.1111/j.1365-2672.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- 32.Stein A, Louveau C, Lepidi H, Ricci F, Baylac P, Davoust B, Raoult D. Q Fever pneumonia: virulence of Coxiella burnetti pathovars in a murine model of aerosol infection. Infect. Immun. 2005;73:2469–2477. doi: 10.1128/IAI.73.4.2469-2477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenwood DP. A relative assessment of putative biological-warfare agents. Massachusetts Institute of Technology Lincoln Laboratory; Boston, MA: 1997. p. 94. (Technical Report 1040). [Google Scholar]
- 34.Rowe-Taitt CA, Golden JP, Feldstein MJ, Cras JJ, Hoffman KE, Ligler FS. Array biosensor for detection of biohazards. Biosens. Bioelectron. 2000;14:785–794. doi: 10.1016/s0956-5663(99)00052-4. [DOI] [PubMed] [Google Scholar]
- 35.Lee WE, Thompson HG, Hall JG, Bader DE. Rapid detection and identification of biological and chemical agents by immunoassay, gene probe assay and enzyme inhibition using a silicon-based biosensor. Biosens. Bioelectron. 2000;14:795–804. doi: 10.1016/s0956-5663(99)00059-7. [DOI] [PubMed] [Google Scholar]
- 36.Halling SM. The effects of magainin 2, cecropin, mastoparan, and melittin on Brucella abortus. Vet. Microbiol. 1996;51:187–92. doi: 10.1016/0378-1135(96)00027-2. [DOI] [PubMed] [Google Scholar]
- 37.Martinez de Tejada G, Pizarro-Cerda J, Moreno E, Moriyon I. The outer membranes of Brucella spp. are resistant to bactericidal cationic peptides. Infect. Immun. 1995;63:3054–3061. doi: 10.1128/iai.63.8.3054-3061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manterola L, Moriyon I, Moreno E, Sola-Landa A, Weiss DS, Koch MHJ, Brandenburg K, Lopez-Goni I. The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J. Bacteriol. 2005;187:5631–5639. doi: 10.1128/JB.187.16.5631-5639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein A, Raoult D. Detection of Coxiella burnetti by DNA amplification using polymerase chain reaction. J. Clin. Microbiol. 1992;30:2462–2466. doi: 10.1128/jcm.30.9.2462-2466.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiele D, Karo M, Krauss H. Monoclonal antibody based capture ELISA/ELIFA for detection of Coxiella burnetii in clinical specimens. Eur J Epidemiol. 1992;8:568–74. doi: 10.1007/BF00146378. [DOI] [PubMed] [Google Scholar]
- 42.Dellacasagrande J, Ghigo E, Hammami SM, Toman R, Raoult D, Capo C, Mege J-L. α(v)β(3) integrin and bacterial lipopolysaccharide are involved in Coxiella burnetti-stimulated production of tumor necrosis factor by human monocytes. Infect Immun. 2000;68:5673–8. doi: 10.1128/iai.68.10.5673-5678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honstettre A, Ghigo E, Moynault A, Capo C, Toman R, Akira S, Takeuchi O, Lepidi H, Raoult D, Mege J-L. Lipopolysaccharide from Coxiella burnetii is involved in bacterial phagocytosis, filamentous actin reorganization, and inflammatory responses through toll-like receptor. J. Immunol. 2004;172:3695–3703. doi: 10.4049/jimmunol.172.6.3695. [DOI] [PubMed] [Google Scholar]
- 44.Aragon AS, Pereira HA, Baca OG. A cationic antimicrobial peptide enhances the infectivity of Coxiella burnetii. Acta Virol. 1995;39:223–226. [PubMed] [Google Scholar]
- 45.Ligler FS, Shriver-Lake LC, Wijesuriya DC. US Patent No. 5,496,700 Optical immunoassay for microbial analytes using non-specific dyes. 1996 March 5;