Abstract
Cdc48p is an abundant and conserved member of the AAA ATPase family of molecular chaperones. Cdc48p performs ubiquitin selective functions, which are mediated by numerous ubiquitin-binding adaptors, including the Npl4p-Ufd1p complex. Previous studies suggest that Cdc48p-containing complexes carry-out many biochemical activities, including ubiquitination, deubiquitination, protein complex segregation and targeting of ubiquitinated substrates to the proteasome. The molecular mechanisms by which Cdc48p containing complexes participate in these processes remain poorly defined. We show here using physiologically relevant Cdc48p substrates (i.e. endoplasmic membrane associated/tethered dimers of Mga2p and Spt23p) and in vitro systems with purified proteins that Cdc48pNpl4p/Ufd1p binds to and promotes segregation of the tethered proteins via a poly-ubiquitin signal present on the membrane-bound proteins. Mobilization does not involve retrotranslocation of the associated anchors. These results provide biochemical evidence that Cdc48pNpl4p/Ufd1p functions as a poly-ubiquitin-selective segregase and that a poly-ubiquitin-Cdc48p-pathway modulates protein interactions at cell membranes.
Introduction
Cdc48p of Saccharomyces cerevisiae is a highly conserved member of the AAA (ATPase Associated with diverse cellular Activities) ATPase family of molecular chaperones and plays an essential role in numerous cellular processes, including membrane fusion, spindle disassembly, ERAD (Endoplasmic Reticulum Associated Degradation), and ubiquitin (Ub)/proteasome-dependent proteolysis (reviewed in Woodman, 2003; Wang et al., 2004; Ye, 2006). Cdc48p harbors two centrally localized ATPase domains (D1 and D2). The protein exists as a hexamer with the D1 and D2 domains forming stacked rings that couple ATP-hydrolysis to conformational changes in the hexamer (reviewed in Pye, 2006). It is postulated that this activity applies mechanical force on substrates, leading to Cdc48p-dependent changes in their conformation.
Although Cdc48p interacts with Ub and poly-Ub chains, Ub-binding adaptors that associate with its amino-terminal domain enhance interactions with ubiquitinated substrates (Meyer et al., 2002; Ye et al., 2003). One of the best-studied Cdc48p adapters is the highly conserved dimeric Npl4p-Ufd1p complex. Studies, performed predominately in yeast, have demonstrated that Npl4p-Ufd1p mediates a subset of Cdc48p activities; functioning in ERAD, mitosis, and activation of membrane-bound transcription factors Mga2p and Spt23p (reviewed in Bays and Hampton, 2002; Wang et al., 2004; Ye, 2006). Multiple domains mediate the Ub binding activity of the complex and there are important differences in both the structure and function of yeast and metazoan Npl4p (Meyer et al., 2002; Ye et al., 2003). Metazoan Npl4p has a carboxy-terminal Ub binding Zinc Finger domain that interacts with free Ub and Lys 48 and Lys 63-linked Ub chains and deletion of this domain abrogates its Ub binding function (Meyer et al., 2002; Ye et al., 2003). Yeast Npl4p lacks this domain and thus substrate binding by the Npl4p-Ufd1p complex is likely mediated by the Ub association function of Ufd1p (Ye et al., 2003). Structural studies have shown that the amino-terminus of Ufd1p has two distinct binding sites for mono-Ub and poly-Ub, although it displays a higher affinity for poly-Ub chains (Park et al., 2005). Also, considering that Ufd1p containing complexes that have been immunopurified from yeast bind Lys 48 linked, but not Lys 63 linked chains, it is probable that substrates containing Lys 48-linked chains are the preferred targets of Cdc48pNpl4p/Ufd1p in yeast (Ye et al., 2003).
Cdc48p complexes have been proposed to function in numerous Ub-related processes in addition to retrotranslocating misfolded ubiquitinated proteins from the ER, including those that have differing consequences (reviewed in Halawani and Latterich, 2006). For example, studies have suggested that Cdc48p complexes recruit E3/E4 ligases to certain substrates and/or modulate their conformation so that they become permissive to ligase-mediated Ub modification (Ishigaki et al., 2004; Yoshida et al., 2005; Richly et al., 2005). In contrast, Cdc48p also interacts with deubiquitinating enzymes and may thus control disassembly of Ub chains from certain substrates (Rumpf and Jentsch, 2006; Mullally et al., 2006). Notwithstanding, our current mechanistic understanding of how Cdc48p participates in these diverse processes is very limited.
As stated above, the Cdc48pNpl4p/Ufd1p complex is required for the activation of the yeast membrane-bound transcription factors Mga2p and Spt23p (Rape et al., 2001; Hitchcock et al., 2001). Mga2p and Spt23p play overlapping roles in the regulation of OLE1 and other lipid metabolism genes (Hoppe et al., 2000; Auld et al., 2006). These proteins are synthesized as 120 kDa (i.e. p120) endoplasmic reticulum (ER)-localized precursors (both proteins contain a carboxy-terminal transmembrane domain) (Hoppe et al., 2000). Upon homo-dimerization via their IPT (Ig-like/Plexins/Transcription factors) domain, one molecule of the dimer undergoes ubiquitination and a proteasome-dependent endo-proteolytic cleavage event, followed by bi-directional degradation (Piwko and Jentsch, 2006). The amino-terminal 90 kDa of the protein (i.e. p90) is spared from complete degradation, presumably due the presence of the highly stable IPT dimerization domain. p90 remains tethered to the membrane via an interaction with unprocessed p120 (Rape et al., 2001; Shcherbik et al., 2003). Previous studies have provided evidence that the Rsp5p Ub ligase and Cdc48pNpl4p/Ufd1p are required for mobilization of the transcriptionally active Mga2p90 and Spt23p90 products from their respective ER bound anchors (Rape et al., 2001; Shcherbik et al., 2003). In terms of Spt23p, Cdc48pNpl4p/Ufd1p-mediated Spt23p90 mobilization has been postulated to be dependent on a Cdc48pNpl4p/Ufd1p interaction with mono-Ub Spt23p90 (Rape et al., 2001). We have previously demonstrated using cell-based systems that nuclear trafficking of non-Ub modified Mga2p90 is associated with poly-ubiquitination of the membrane anchor and is suppressed in yeast strains harboring mutations in rsp5, cdc48 or npl4 (Shcherbik et al., 2003). In this study, we have exploited in vitro systems to more precisely define physical and functional interactions between Rsp5p, Cdc48pNpl4p/Ufd1p and heteromeric (i.e. p120-p90) Mga2p and Spt23p complexes.
Results
Cdc48pGSTNpl4p/Ufd1p binding to an Mga2p120-Mga2p90 complex requires Rsp5p-dependent poly-ubiquitination in vitro
Because our past cell-based studies focused predominately on Mga2p, we first set out to determine using purified components if an interaction between Cdc48pNpl4p/Ufd1p and an Mga2p120-Mga2p90 complex is direct and requires ubiquitination of Mga2p120. Recombinant GST-tagged Cdc48p, Npl4p and Ufd1p were produced in bacteria and captured on glutathione sepharose. Cdc48p and Ufd1p were liberated from the solid support with Procession Protease. Proteins were mixed with glutathione beads containing GST-Npl4p and complexes were allowed to form. A fraction of this material was pelleted and analyzed by SDS-PAGE and Coomassie blue staining to verify Cdc48pGSTNpl4p/Ufd1p complex formation. As shown in the left panel of Fig 1A, Cdc48pGSTNpl4p/Ufd1p preparations contained stoichiometric amounts of Cdc48p, GST-Npl4p and Ufd1p. We were unable to detect Cdc48p and Ufd1p on beads containing GST alone.
Fig 1. Cdc48pGSTNpl4p/Ufd1p interacts with poly-ubiquitinated Mga2p120 and Mga2p90.

(A) (Left panel) Recombinant GST-tagged Cdc48p, Npl4p and Ufd1p proteins were produced in bacteria and purified with glutathione beads. GST-tagged Cdc48p and Ufd1p were liberated from beads using Procession Protease. Proteins were then mixed with bead-bound GST-Npl4p or GST alone control and protein complexes were allowed to form. Beads were pelleted, resuspended in SDS-PAGE loading buffer, boiled, resolved by SDS-PAGE, and the formation of Cdc48pGSTNpl4p/Ufd1p complex was verified by Coomassie blue staining. (Right Panel) Microsomes were isolated from yeast expressing FLAG-tagged Mga2p or empty vector control and solublized. Proteins were immunoprecipitated with anti-FLAG antibody conjugate agarose. Proteins were eluted with FLAG peptide, separated by SDS-PAGE, and visualized by Coomassie blue staining. (B) Agarose captured Mga2p120-Mga2p90 complexes were placed in in vitro ubiquitination reactions containing indicated E1, the E2 Ubc1p, and varying amounts of Rsp5p (600 ng, 200 ng, 66 ng). After termination, agarose beads were washed to remove enzymes and complexes were eluted with FLAG peptide. Western blotting was performed on duplicate gels with anti-FLAG and anti-Ub antibodies. * Denotes an alternatively process Mga2p product, see text for details. (C) Products from the in vitro ubiquitination reactions were incubated with glutathione beads containing GST or Cdc48pGSTNpl4p/Ufd1p. Beads were washed, resuspended in SDS-PAGE loading buffer and samples were boiled. Equal amounts of eluted material were run on duplicate 8.0% SDS-polyacrylamide gels and western blotting was carried-out separately with anti-FLAG and anti-Ub antibodies. Ponceau S staining of both membranes (only the one for the anti-FLAG blot is presented) was performed to show equivalent amount of recombinant proteins in each binding reaction. (D) Binding assays were carried-out using the indicated GST-fused proteins/complexes and ubiquitin modified or unmodified Mga2p120-Mga2p90 complexes.
Since we encountered numerous technical problems generating dimeric Mga2p120-Mga2p90 using recombinant proteins prepared from bacteria, we immunopurified Mga2p120-Mga2p90 complexes from yeast microsomes. Microsomes were prepared from cells overexpressing an amino-terminal FLAG-tagged Mga2p. Membranes were solublized using Triton X-100 containing buffer and complexes were captured onto anti-FLAG antibody conjugated agarose beads. As shown in the right panel of Fig 1A, this approach generated pure (greater than 90%) Mga2p120-Mga2p90 complex. The approximate 100 kDa band of lower abundance (denoted by a * in Figs 1A and 1B) that is detected here as well as in other figures is anti-FLAG reactive and anti-Ub non-reactive protein. This Mga2p product likely represents based on the data presented in Fig 1A and 1B, our interaction studies (see below) and recently published data (Piwko and Jentsch, 2006) an alternative proteasome generated Mga2p120-tethered Mga2p protein that lacks the carboxy-terminal transmembrane and Rsp5p binding site.
Bead coupled Mga2p120-Mga2p90 was next placed in in vitro ubiquitination reactions containing or lacking the recombinant E3 Ub ligase Rsp5p. We have shown previously that Mga2p is a direct target of Rsp5p and Rsp5p-Mga2p binding is mediated by an imperfect WW interacting motif (i.e. LPKY) that is located within the carboxy-terminal domain of Mga2p120 (Shcherbik et al., 2004). After completion of the ubiquitination reactions, beads were washed and Mga2p was eluted with FLAG peptide. Eluted products were analyzed by western blotting using anti-FLAG and anti-Ub antibodies. As shown in Fig 1B, high molecular weight ubiquitinated Mga2p products were clearly evident in reactions containing E1, E2, and Rsp5p enzymes, but not in reactions supplemented with only E1 and E2 proteins. In addition, we did not detect a size shift in the molecular weight of Mga2p90 in any ubiquitination reactions. It also deserved to be noted that the appearance of ubiquitinated products correlates with a decrease in Mga2p120 and not Mga2p90. These results indicate that the preferred Rsp5p target in an Mga2p120-Mga2p90 complex is Mga2p120.
Binding assays were carried out with glutathione bead-coupled Cdc48pGSTNpl4p/Ufd1p and Mga2p products that were generated as described above. As shown in Fig 1C, we were only able to detect a strong interaction between Cdc48pGSTNpl4p/Ufd1p and Mga2p90 using inputs derived from ubiquitination reactions containing E1, E2 and Rsp5p enzymes. Also, in addition to pulling-down unmodified Mga2p90 in the binding reactions, we also pelleted high molecular weight Ub modified Mga2p proteins (Fig 1C). Ponceau S staining of blots prior to probing with the indicated antibodies shows equivalent amount of Cdc48pGSTNpl4p/Ufd1p in all pull-downs. Similar results were obtained with a Cdc48pNpl4p/GSTUfd1p complex, where GST-Ufd1p was used in the preparations (Fig 1S of supplemental data). It also deserves to be noted that we routinely pellet approximately 20% of substrate input (i.e. Ub-modified Mga2p120-Mga2p90 complex) with the optimal amount of Cdc48pGSTNpl4p/Ufd1p or Cdc48pNpl4p/GSTUfd1p placed in the binding assays. Besides performing interaction assays with Cdc48pGSTNpl4p/Ufd1p, we also carried-out binding studies with GST-Cdc48p and GST-Npl4p/Ufd1p complex to determine if interaction is mediated by the AAA ATPase and/or its dimeric co-factor. As depicted in Fig 1D, interaction with poly-ubiquitinated Mga2p120 (Poly-UbMga2p120) and Mga2p90 was clearly evident when the Ub-binding adapter complex Npl4p/Ufd1p was present in the binding reactions. Curiously, we were only able to pull-down a small percentage of Poly-UbMga2p120-Mga2p90 in reactions containing GST-Cdc48p (a weak ubiquitin signal is apparent on long exposures). Considering that previous studies have shown that Cdc48p has affinity for poly-ubiquitinated substrates when using a smaller tag on the protein or coupling untagged Cdc48p to a GST-fusion harboring the Cdc48p interaction domain of Ufd1p (Meyer et al., 2002 and Ye et al., 2003), it is likely that the GST tag on GST-Cdc48p negatively affects Cdc48p binding.
Considering that the immunopurified Mga2p120-Mga2p90 complex is not 100% pure, it is possible that Rsp5p target(s) other than Mga2p120 mediates an interaction with Mga2p90. To address this possibility, we performed additional binding assays with two different Mga2p mutants, one that contains only the 90 kDa amino terminal portion of Mga2p (this mutant was immunopurified from total cell extract since it lacks the transmembrane domain and is not ER-bound) and another (i.e. Mga2pΔLPKY) that is missing 4 amino acids corresponding to the Rsp5p binding motif (Shcherbik et al., 2004). Fig 2 shows that Cdc48pGSTNpl4p/Ufd1p is unable to interact with Mga2p90 that is derived from ubiquitination reactions harboring only Mga2p90 or a Mga2p120ΔLPKY-Mga2p90 complex that lacks the Rsp5p binding site. These results provide strong evidence that a Cdc48pGSTNpl4p/Ufd1p interaction with Mga2p90 is mediated via poly-ubiquitinated Mga2p120.
Fig 2. Cdc48pGSTNpl4p/Ufd1p interaction with Mga2p90 is dependent on the presence of Mga2p120 and the Rsp5p binding motif.
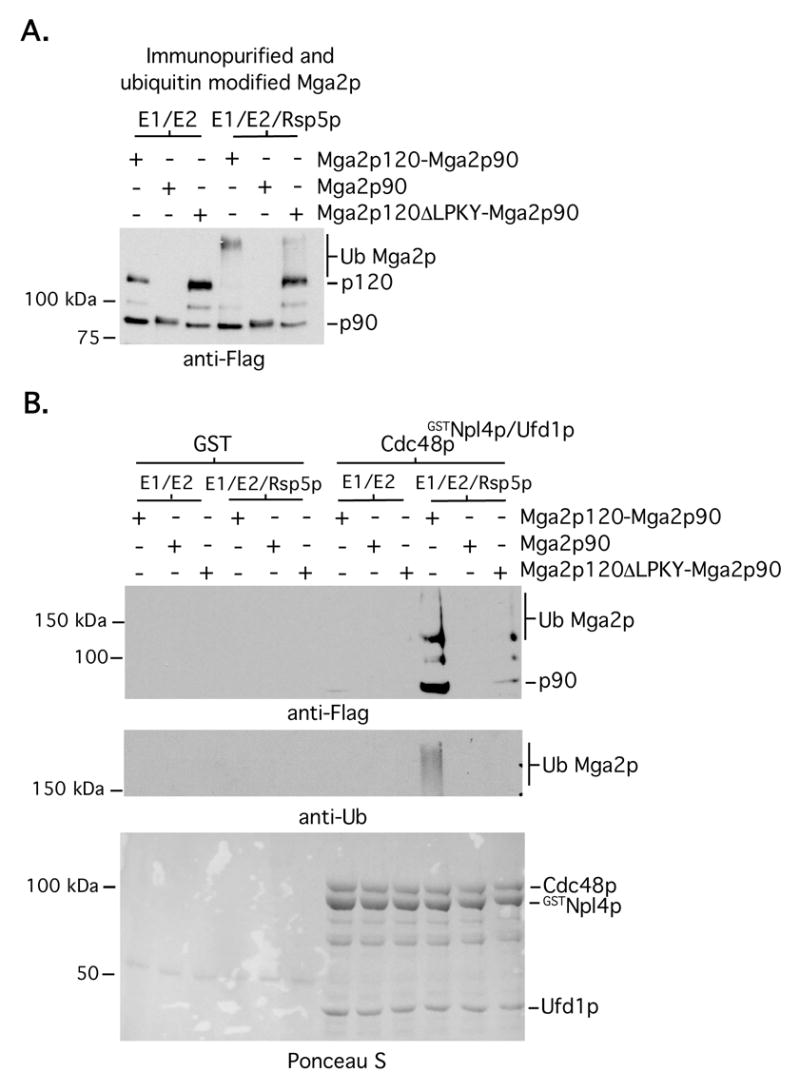
Inputs (A) were prepared and binding assays (B) were performed as in Fig 1, except that immunopurified FLAG-tagged Mga2p90 (prepared from whole cell extracts of cells harboring a galactose-inducible FLAG-tagged mga2 truncation mutant) and FLAG-tagged Mga2p120ΔLPKY-Mga2p90 complexes were also included.
We next tested if Cdc48pGSTNpl4p/Ufd1p binding to Mga2p120-Mga2p90 requires poly-Ub chain formation on the substrate. Ubiquitination reactions were performed with WT (wild-type) Ub or a chemically modified Ub mutant (i.e. methylated Ub) that is unable to form chains. Products from these reactions were then used in Cdc48pGSTNpl4p/Ufd1p pull-down assays. As shown in Fig 3A, we were unable to detect an interaction between Mga2p120-Mga2p90 and Cdc48pGSTNpl4p/Ufd1p using substrate generated in ubiquitination reactions containing methylated Ub. One possibility however is that Cdc48pGSTNpl4p/Ufd1p is defective at binding to methylated Ub. To address a preference of Cdc48pGSTNpl4p/Ufd1p for interacting with Poly-UbMga2p120-Mga2p90 by another approach, we first modified the Mga2p120-Mga2p90 complex with mutant Ub (i.e. K48R/K63R) harboring substitutions at Lys 48 and Lys 63. Rsp5p is unable to efficiently promote poly-Ub chain assembly when ubiquitination reactions are performed with this mutant and reduced amounts of Rsp5p and the predominant products formed are multiple-mono-ubiquitinated Mga2p120 (left panel of Fig 3B). As shown in the right panel of Fig 3B, we were also unable to detect an interaction between Cdc48pGSTNpl4p/Ufd1p and Mga2p120-Mga2p90 that had been modified with the K48R/K63R Ub mutant. We conclude from these studies that Cdc48pGSTNpl4p/Ufd1p interacts with an Mga2p120-Mga2p90 complex via an Rsp5p-dependent poly-ubiquitin signal that is placed on the membrane interacting anchor. We also deduce based on previous studies showing that yeast Cdc48pNpl4p/Ufd1p does not interact with Lys 63-linked chains (Ye et al., 2003) that Lys-48 linked chains that are present on Mga2p120 mediate a Cdc48pGSTNpl4p/Ufd1p-Mga2p120-Mga2p90 interaction.
Fig 3. A Cdc48pGSTNpl4p/Ufd1p-Mga2p120-Mga2p90 interaction requires poly-ubiquitination.
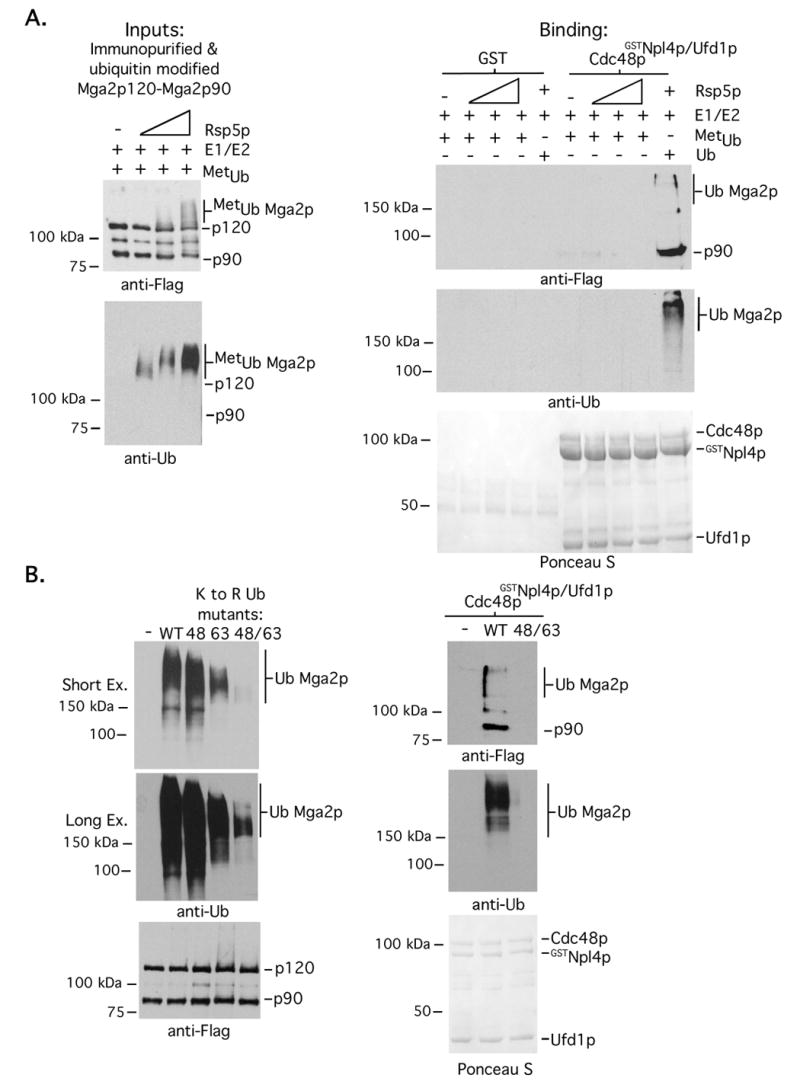
(A). (Left panel) Mga2p120-Mga2p90 complexes were prepared and immobilized on anti-FLAG antibody conjugated agarose beads. Ubiquitination reactions were carried out in the presence of methylated Ub (metUb), E1, the E2 Ubc1p, and varying amounts (30 ng, 100 ng and 300 ng) of Rsp5p. Reaction products were eluted from beads with FLAG peptide and analyzed by western blotting in duplicate with indicated antibodies. (Right panel) GST pull-downs were performed as described in the legend of Fig 1C. (B) (Left panel) Ubiquitination reactions were carried out as described in A in the presence of WT or the indicated ubiquitin mutants and 70 ng of Rsp5p. Reaction products were eluted from beads with FLAG peptide and analyzed by western blotting in duplicate with indicated antibodies. (Right panel) GST pull-downs were performed with WT and K48R/K63R ubiquitin modified Mga2p120-Mga2p90 complexes as described in the legend of Fig 1C.
Cdc48pNpl4p/Ufd1p promotes Mga2p90 mobilization from membranes and segregation of Mga2p90 from bead captured Poly-UbMga2p120
In addition to the above presented interaction experiments, we also examined how recombinant Cdc48pNpl4p/Ufd1p affects mobilization of Mga2p proteins from the ER membrane using a cell free system. Microsomes were prepared from cells expressing epitope-tagged Mga2p. Recombinant GST-fused Cdc48p, Npl4p and Ufd1p proteins (alone and in complexes) were generated as described above, except that Procession Protease was used to liberate proteins and complexes from glutathione beads (the purity of these as judged by SDS-PAGE and Coomassie blue staining are shown in the left panel of Fig 4A). Proteins, ATP, and an ATP regeneration system were added to microsomes and samples were incubated at 27°C for 1 hour. Samples were then subjected to ultracentrifugation and the amounts of Mga2p120 and Mga2p90 in the supernatant and pelleted membranes were determined by western blotting. As shown in Fig 4A (right panel), the greatest amount of soluble Mga2p90 was present in the reaction harboring Cdc48pNpl4p/Ufd1p and inclusion of the ATP depleting enzyme Apyrase suppressed Cdc48pNpl4p/Ufd1p-mediated Mga2p90 release from membranes. Interestingly, we could not detect the presence of Mga2p120 or high molecular weight modified Mga2p120 peptides, indicating that at least in vitro, Mga2p90 mobilization is not coupled to Cdc48pNpl4p/Ufd1p-induced retrotranslocation of the membrane-bound anchor (Fig 4A). A slight increase in soluble Mga2p90 was also detected in reactions harboring an Npl4p/Ufd1p complex, presumably due to the present of endogenous membrane-associated Cdc48p (Rape et al., 2001; Hitchcock et al., 2001).
Fig 4. Cdc48pNpl4p/Ufd1p-dependent Mga2p90 mobilization requires Rsp5p and the mobilized product is not Ub modified.
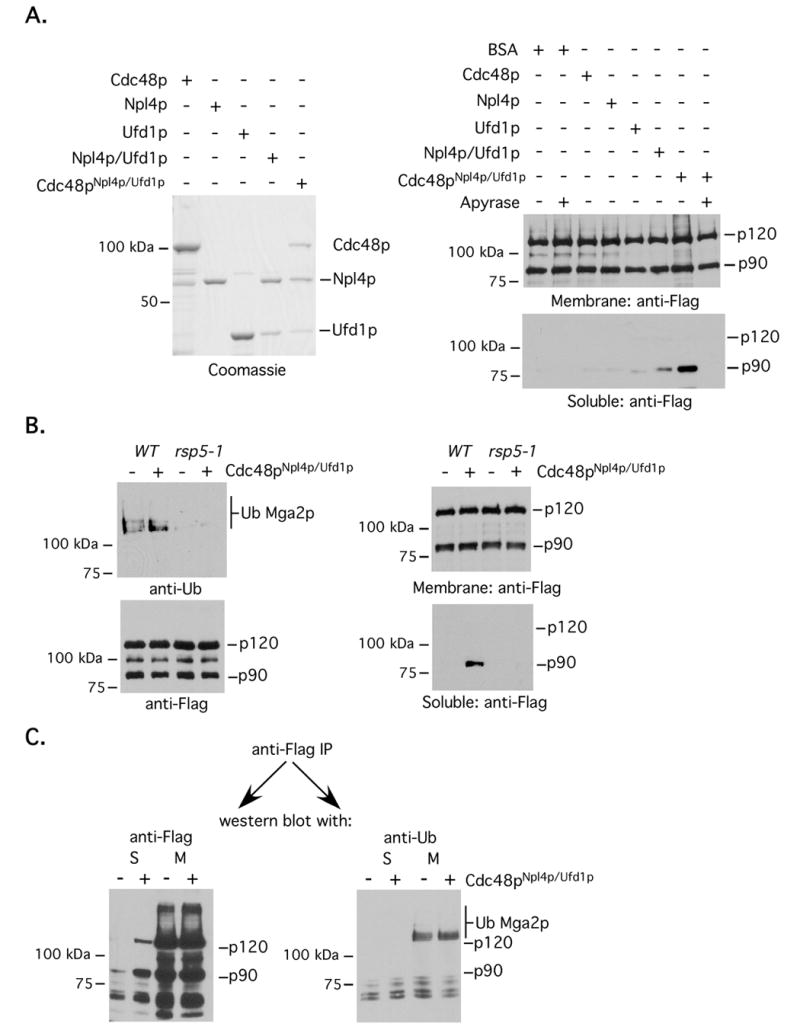
(A) (Left panel) Recombinant GST fused proteins and complexes were prepared and captured on glutathione sepharose. Products were remove from beads with Precision Protease, resolved by SDS-PAGE and visualized by Coomassie blue staining. (Right panel) Microsomes were isolated from cells expressing amino-terminal FLAG-tagged Mga2p. In vitro mobilization assays were setup with microsomes and the indicated recombinant proteins, complexes, ATP, ATP energy regeneration system and/or Apyrase. After reactions were complete, samples were separated into insoluble (membrane) and soluble fractions by ultracentrifugation and the amount of Mga2p120 and Mga2p90 in these fractions were determined by western blotting. (B) (Left panels) Microsomes were prepared from WT or rsp5-1 cells expressing FLAG-tagged Mga2p at the non-permissive temperature. Membranes were solublized and immunoprecipitations were performed with an anti-FLAG antibody. Immunoprecipitated proteins were resolved by SDS-PAGE, transferred to nitrocellulose membrane and probed with indicated antibodies. (Right panel) In vitro mobilization assays were carried-out using microsomes described in the left panel. (C) After termination of in vitro mobilization reactions, anti-FLAG immunoprecipitations were performed using material from the supernatant (S) as well as the solublized pellet (M). SDS-PAGE and western blotting were then carried-out with anti-FLAG and anti-Ub antibodies.
To determine whether Cdc48pNpl4p/Ufd1p-mediated Mga2p90 mobilization is dependent on an Rsp5p-generated signal, we performed mobilization assays using microsomes isolated from rsp5-1 cells grown at 37°C. rsp5-1 cells harbor a temperature sensitive rsp5 gene mutation and the ligase is not functional when cells are grown at 37°C. Fig 4B shows that the level of ubiquitinated Mga2p, but not total Mga2p, is reduced in microsomes prepared from rsp5-1 mutant cells (left panel) at the non-permissive temperature. The right panel of Fig 4B shows that Cdc48pNpl4p/Ufd1p is unable to promote Mga2p90 release (right panel) from rsp5-1 derived microsomes. Because Cdc48pNpl4p/Ufd1p-mediated mobilization of homologous Spt23p90 from membranes appears to be dependent on an interaction with mono-ubiquitinated Spt23p90 (Rape et al., 2001), we wanted to investigate if soluble Mga2p90 has such a modification. As shown in Fig 4C, we could not any ubiquitin modified Mga2p, including mono-ubiquitinated Mga2p90, in the soluble fraction using an antibody that recognizes both a single attached Ub moiety or assembled Ub chains. Also, we have not noticed a difference in the relative migration of soluble versus membrane-bound Mga2p90, even when the proteins are resolved on large gels (data not shown).
Although the above-presented findings point to a direct Mga2p120-Mga2p90 segregase function of Cdc48pNpl4p/Ufd1p in a biologically relevant in vitro mobilization system, it is entirely possible based on the nature of the system that the disassociation activity is indirect. To address these issues, we set out to establish a microsome-free in vitro segregation assay. Epitope tagged Mga2p120-Mga2p90 complexes were prepared and Ub modified as described previously, except that a MGA2 expression construct harboring both amino (i.e. FLAG) and carboxy-terminal (i.e. HA) tags was used as the substrate. After release with FLAG peptide, Mga2p120-Mga2p90 complexes were recaptured on anti-HA antibody conjugated beads to give Mga2p90 that is anchored to a solid support via the carboxy-terminal epitope tag that is present on associated Mga2p120. This complex was then placed in in vitro segregation assays with the indicated proteins and complexes, ATP and an ATP energy regeneration system. Beads were pelleted and the amount of Mga2p90 released from the solid support was determined by western blotting with the anti-FLAG antibody. As shown in Fig 5, Cdc48pNpl4p/Ufd1p induces liberation of Mga2p90 from beads. This activity was ATP dependent since segregation was not observed in reactions containing Apyrase and it also required Rsp5p-dependent ubiquitination. We also noticed that Cdc48p alone (no GST tag) promoted ligase-dependent Poly-UbMga2p120-Mga2p90 segregation, although not to the same extent as with the entire complex. This result suggests that at least under the conditions used here, Cdc48p has sufficient ubiquitin binding activity and is consistent with the idea that both Cdc48p and Npl4p/Ufd1p have Ub interaction domains (Meyer et al., 2002; Ye et al., 2003). Alternatively, these preparations may contain co-purifying Npl4p and Ufd1p that is able to function with recombinant Cdc48p. In the absence of available antibodies recognizing yeast Npl4p and Ufd1p, we have address the later possibility in two ways. First, we have analyzed inputs for the in vitro segregation assays via Coomassie blue staining and have not found that proteins of similar molecular weight as Npl4p or Ufd1p co-purify with Mga2p (Fig2SA). In addition, Fig 2SB show that Mga2p90 release in the in vitro segregation assay plateaus at a relatively high amount of input Cdc48p, a result that is inconsistent with the notion that Cdc48p-mediated Mga2p90 segregation is dependent on a potentially small/sub-stoichiometric quantity of contaminating Npl4p/Ufd1p. We thus conclude that Cdc48p dependent Mga2p90 segregation in this system is direct and is enhanced by the presence of Npl4p and Ufd1p.
Fig 5. Cdc48pNpl4p/Ufd1p promotes Mga2p90 segregation from bead-bound Ub modified Mga2p120.

Mga2p120-Mga2p90 complexes were prepared, Ub modified and eluted from beads with anti-FLAG peptide as described before, except that cells expressing Mga2p harboring both amino (FLAG) and carboxy (HA) terminal tags were used. After elution with FLAG peptide, Ub-modified material was re-captured onto another solid support with anti-HA antibody conjugated agarose. Fifty ng of material was placed into reactions containing ATP, ATP energy regeneration system and 100 ng of BSA, indicated recombinant proteins, complexes (100 ng in total), and/or Apyrase. Beads were pelleted and the amount of bead-bound and released Mga2p was determined by western blotting.
Cdc48pNpl4p/Ufd1p binds and segregates heterodimeric Poly-UbSpt23p120-Spt23p90 complexes
As mentioned in the introduction, Cdc48pNpl4p/Ufd1p-dependent segregation of Spt23p90 from tethered Spt23p120 has been proposed to be via an Spt23p90 mono-ubiquitin signal that is a remnant of the proteasome-dependent processing reaction (Rape et al., 2001). Curiously, we could not detect any mono-ubiquitinated Spt23p90 when we immunopurified FLAG-tagged Spt23p120-Spt23p90 complexes from microsomes and eluted the immunoprecipitated proteins with FLAG peptide (Fig 6A). Only large molecular weight ubiquitin modified products greater than 120 kDa were observed (Fig 6A). To determine if Cdc48pNpl4p/Ufd1p binds and segregates Spt23p120-Spt23p90 via a mechanism similar to that of Mga2p, we employed analogous approaches and experiments as presented above using immunopurified Spt23p120-Sp23p90 complexes. Similar to our results with Mga2p, we were only able to detect an interaction between Cdc48pNpl4p/GSTUfd1p and both Ub modified Spt23p120 and non-Ub modified Spt23p90 when the input was derived from ubiquitination reactions containing Rsp5p and WT Ub (Fig 6B). Mobilization and segregation assays were also performed using microsomes and bead-bound Spt23p120-Spt23p90, respectively. As shown in Fig 7A, Cdc48pNpl4p/Ufd1p promotes mobilization of Spt23p90 from microsomes and the immunopurified Spt23p90 product from the soluble fraction is not ubiquitin modified. Finally, Cdc48p and to a greater extent, Cdc48pNpl4p/Ufd1p, facilitates release of Spt23p90 from its bead-bound Poly-UbSpt23p120 partner (Fig 7B). We conclude from these experiments that Cdc48pNpl4p/Ufd1p promotes un-tethering of Mga2p90 and Spt23p90 from their respective Poly-Ub modified membrane-bound anchors via a similar mechanism.
Fig 6. Cdc48pNpl4p/GSTUfd1p binds heterodimeric Spt23p120-Spt23p90 via an Spt23p120 poly-ubiquitin signal.
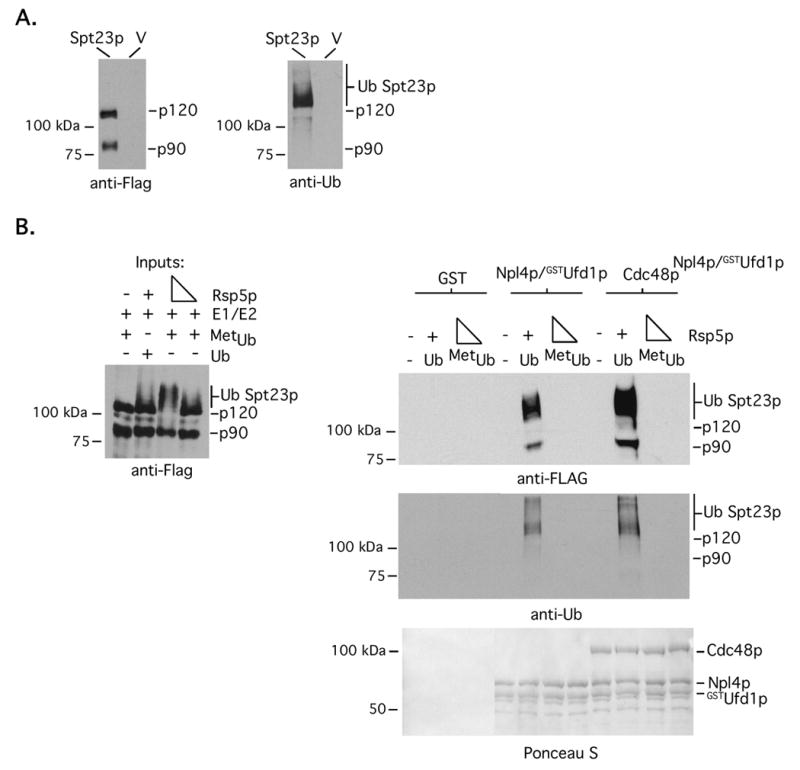
(A) Microsomes were prepared from cells harboring galactose-inducible FLAG-tagged SPT23 or empty vector (V), solublized and immunoprecipitations were performed with anti-FLAG antibody conjugated agarose. After extensive washing, Spt23p was eluted with FLAG peptide. Eluted material was separated by SDS-PAGE and western blotting was performed with anti-FLAG and anti-Ub antibodies. (B) In vitro binding assays with the indicated complexes were performed with Ub modified Spt23p120-Spt23p90 substrates as described previously for Mga2p.
Fig 7. Cdc48pNpl4p/Ufd1p promotes Spt23p mobilization from microsomes and segregation of Poly-UbSpt23p120-Spt23p90 complexes in vitro.
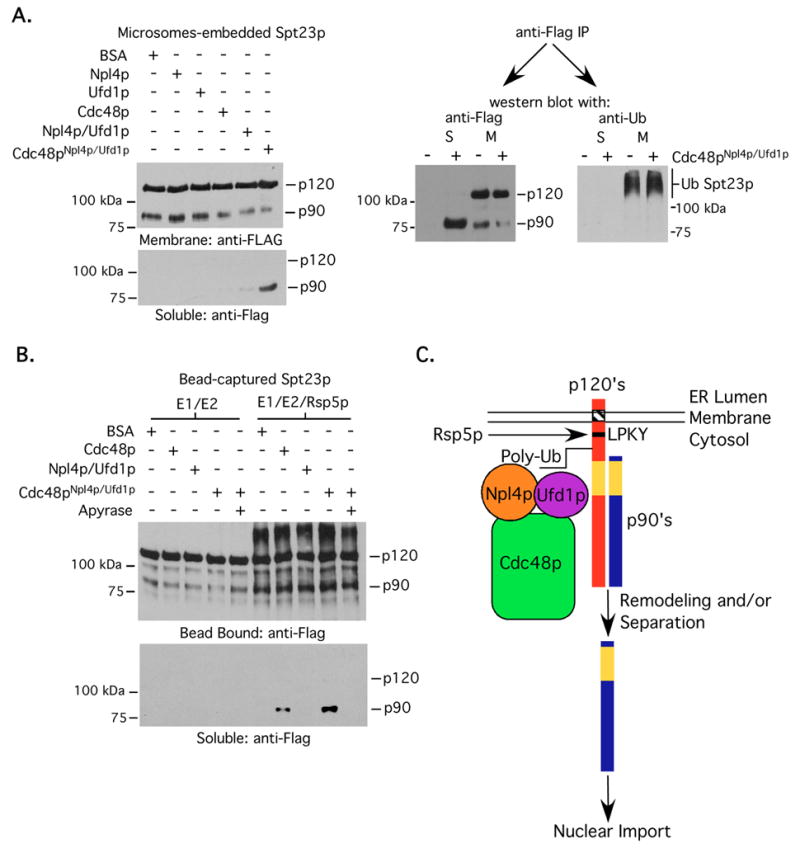
Microsome-based mobilization and analysis of mobilized products (A), and bead-based segregation (B) studies were performed as described in the above figure legends for Mga2p, except that epitope-tagged Spt23p proteins carrying the identical tags as Mga2p were used for the studies. (C) Proposed model of Rsp5p-Cdc48pNpl4p/Ufd1p-mediated membrane-bound transcription factor mobilization. Sites of ubiquitin conjugation remained to be identified.
Discussion
We previously proposed a model based on cell-based studies where Cdc48pNpl4p/Ufd1p promotes liberation of Mga2p90 from the ER membrane via an interaction with poly-ubiquitinated Mga2p120 (Shcherbik et al., 2003). Because mobilization was associated with an increase in poly-ubiquitinated Mga2p120 in cells lacking Cdc48p or Npl4p function, we initially thought that Mga2p90 release/disassociation is coupled to Mga2p120 retrotranslocation and proteasome-dependent degradation of the membrane bound anchor. The in vitro binding data presented here supports the “signal” component of our model, showing that the interaction between Cdc48pNpl4p/Ufd1p and an Mga2p120-Mga2p90 is direct and mediated by an Rsp5p-induced poly-Ub signal on Mga2p120 (see Fig 7C for a depiction of our model). However, our in vitro mobilization/segregation data suggests that Cdc48pNpl4p/Ufd1p-dependent release of Mga2p90 is not coupled to retrotranslocation of Mga2p120 and the segregase function of Cdc48pNpl4p/Ufd1p is sufficient for un-tethering. Although we can’t rule out the possibility that our in vitro assays are lacking components needed for retrotranslocation, we have not observed significant differences in Mga2p120 half-life in WT versus cdc48 or npl4 mutant strains (NS and DSH, unpublished data). Thus, it is likely that Cdc48pNpl4p/Ufd1p promotes Mga2p90 liberation from membranes both in vitro and in vivo by simply segregating tethered Mga2p90 from its poly-Ub labeled membrane-bound anchor. If this is the case, why do ubiquitinated forms of Mga2p120 accumulate in cells lacking a functional Cdc48pNpl4p/Ufd1p complex? In light of recent discoveries showing that this complex also associates with deubiquitinating enzymes (Rumpf and Jentsch, 2006), it is conceivable that Cdc48pNpl4p/Ufd1p-dependent mobilization of Mga2p90 is coupled with Cdc48pNpl4p/Ufd1p mediated deubiquitination of Mga2p120 and this could serve as a recycling mechanism to generate unmodified Mga2p120 monomers. Alternatively, these forms may represent a small amount of misfolded non-functional poly-ubiquitinated Mga2p120 that is normally cleared by the ERAD pathway. Notwithstanding, further experiments are needed to determine the mechanism responsible for accumulation of poly-ubiquitinated Mga2p120 in cells lacking Cdc48pNpl4p/Ufd1p function.
Previous studies have shown that Cdc48pNpl4p/Ufd1p also plays a role in promoting mobilization of Spt23p90 from the ER to the nucleus in cells and release of this protein into the soluble fraction from microsomes in vitro (Rape et al., 2001). Consistent with these results, we have found that Cdc48pNpl4p/Ufd1p induces release of both Mga2p90 and Spt23p90 into the soluble fraction without promoting retrotranslocation of the entire complex. Thus, the major disconnect between the proposed models is the nature of the ubiquitin signal responsible for promoting a Cdc48pNpl4p/Ufd1p interaction. Previous studies with Spt23p have proposed that mono-ubiquitination of Spt23p90 is the signal (Rape et al., 2001). This conclusion is based on immunoprecipitating Spt23p from solublized microsomes and performing an anti-Ub western blot with the bead bound material. Using an additional purification step (i.e. elution of immunopurified FLAG tagged Spt23p120-Spt23p90 complexes from agarose beads with a FLAG peptide), we have only detected ubiquitinated Spt23p that is of a greater molecular weight than 120 kDa and we have not been able to identify an ubiquitinated 90 kDa product. In addition, we have found using the microsome-based in vitro mobilization assay that the Cdc48pNpl4p/Ufd1p mobilized Spt23p product is not ubiquitin modified and an interaction between Cdc48pNpl4p/Ufd1p and Spt23p120-Spt23p90 complexes requires Spt23p120 poly-ubiquitination. We therefore propose that Cdc48pNpl4p/Ufd1p promotes liberation of Spt23p90 and Mga2p90 from the ER by a similar mechanism.
Although it is clear from our in vitro studies presented here that Cdc48pNpl4p/Ufd1p directly mediates the separation of Mga2p120-Mga2p90 and Spt23p120-Spt23p90 complexes via a poly-ubiquitination signal present on the membrane-bound anchors, the mechanism by which it does so requires further investigation. It is possible that the Cdc48pNpl4p/Ufd1p complex breaks non-covalent interactions between the dimerized proteins or remodels a domain (i.e. the dimerization domain for example) of one of the proteins so that it looses the ability to dimerize. Also, it remains unclear if Cdc48p works in cis on the ubiquitinated anchor or in trans on unmodified Mga2p90. Further experiments, including structural-based studies, will be required to address these precise mechanisms. Also of interest will be to define if and how the Cdc48pNpl4p/Ufd1p-dependent process is regulated and how the various Ub binding domains present in this complex contribute to segregation. Mga2p and Spt23p are embedded in membranes and they regulate genes involved in lipid metabolism (Hoppe et al., 2000, Auld et al., 2006). It is therefore conceivable that Cdc48pNpl4p/Ufd1p-dependent Mga2p90 and/or Spt23p90 release is coupled to membrane-fluidity sensing mechanisms. In fact, the percentage of p90 products that are mobilized from membrane or beads in our in vitro assays are admittedly low (approximately 10%), perhaps pointing to other important cooperating events such as additional modifications of the membrane-bound transcription factors or members of Cdc48pNpl4p/Ufd1p complex. As for the various Ub interaction domains (likely present on at least Cdc48p and Ufd1p), it will be interesting to determine their relative contributions to the interaction and segregation process. It is conceivable that multiple interactions between the poly-Ub signals present on Mga2p/Spt23p and the various Ub binding domains play a positioning role so that only a specific domain of the protein is destabilized. It is also possible that positioning, length and perhaps type of ubiquitin linkage will play critical roles in Cdc48pNpl4p/Ufd1p-mediated segregation.
As stated in the introduction, results of previous cell based studies have suggested that abundant and highly conserved Cdc48p containing complexes function in numerous processes, including retrotranslocation of mis-folded ER protein, segregation, ubiquitination, and deubiquitination. Studies presented here with heteromeric transcription factor complexes provide for the first time direct biochemical evidence to support a segregase function for Cdc48p. Importantly, this Cdc48p-dependent segregase function plays a critical step in their activation and does so by a mechanism that is not tied to the proteasome. Cdc48p segregase function may however be important for proteasome dependent events as well, such as separating Ub modified from non-modified binding partners so that only the ubiquitinated ones are targeted to the proteasome for destruction. Considering the large number of membrane-localized proteins that undergo poly-ubiquitination, it will be of interest for future studies to determine if Cdc48p (or p97/VCP in mammalian cells) plays generalized role in regulating the sub-cellular localization of membrane-tethered proteins or protein interactions at membranes via a proteasome independent mechanism. Also, bearing in mind the relative abundance of Cdc48p in cells and the growing number of potential Cdc48p adapters (reviewed in Elsasser and Finley, 2005), it will not be surprising if Cdc48p dependent segregase activity is important in the regulation of a large percentage of eukaryotic proteins.
Experimental Procedures
Yeast strains and plasmids
See supplementary information for a description of yeast strains and plasmids used in this study.
Substrate generation
INVSc cells containing galactose-inducible epitope-tagged MGA2 or SPT23 were grown to saturation in glucose media and then placed in galactose media overnight. Cells were harvested and microsomes were purified as described previously (Shcherbik et al., 2003). Microsomes were solublized in SLIP buffer (150 mM NaCl, 50 mM HEPES pH 7.5, 10% glycerol, 0.1% triton-X 100, 10 μM proteasome inhibitor MG115, and protease cocktail). FLAG-tagged Mga2p/Spt23p proteins were captured on anti-FLAG M2 antibody coupled agarose (Sigma) overnight at 4 C. Beads were washed 3 times with high-salt-RIPA buffer (50mM Tris HCl pH 8.0, 500 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS) and 2 times with 25 mM Tris HCl pH 7.5. In vitro ubiquitination reactions with bead bound material (contains approximately 1 μg of protein per reaction) and Rsp5p were performed as described previously (Shcherbik et al., 2004) using yeast E1 (Boston Biochem) and the yeast E2 Ubc1p. Recombinant WT and mutant Ub used in the various reactions were purchased from Boston Biochem. After reactions were complete, beads were washed 2 times with high salt RIPA buffer and 3 times with elution buffer (50mM Tris HCl pH 8.0, 200 mM KCl, 2.5 mM MgCl2, 1% triton X-100, 5% glycerol, 1mM PMSF, 10 μM MG115, 0.1mM DTT, and protease inhibitor cocktail) and FLAG-tagged Mga2p/Spt23p proteins were specifically eluted with 100 ng/μl of FLAG peptide (Sigma). To assess ubiquitination status of substrate protein, supernatants were collected, resolved by SDS-PAGE, transferred to nitrocellulose membrane and subjected to western blotting using M5 anti-FLAG antibody (Sigma) and anti-Ub antibody P4D1 (Santa Cruz Biotechnology).
Protein complex formation
Individual recombinant proteins were expressed and purified from bacteria as described in supplemental material. Glutathione sepharose (GE Healthcare/Amersham) containing approximately 10 μg of GST-coupled Npl4p was first equilibrated with WB buffer and incubated with 20 μg of recombinant Cdc48p and/or Ufd1p. Proteins were incubated overnight at 4°C. Beads were washed in WB buffer (50 mM Tris HCl pH 8.0, 200 mM KCl, 2.5 mM MgCl2, 1 mM ATP, 5% glycerol, 0.2% Triton X-100, 1μM ZnCl2, supplemented with 1 mM PMSF, 10 μM MG115, 0.1 mM DTT, and protease inhibitor cocktail) three times, and resuspended in the same buffer, supplemented with protease inhibitors and PMSF. To remove complexes from the beads, samples were subjected to Precision Protease (GE Healthcare/Amersham) treatment. Complexes containing GST-tagged Ufd1p were prepared in the same manner, except that GST-coupled Ufd1p was mixed with Precision Protease-liberated Npl4p and Cdc48p.
GST-pull down assay
Equal amounts of GST-fused proteins (1 μg) were mixed with purified and modified Mga2p/Spt23p substrates (100 ng). Binding reactions were carried out in 200μl of WB buffer for 4 hrs at 4°C. Beads were washed 3 timed with the WB buffer supplemented with 1% Triton X-100 and proteins were eluted with SDS-PAGE loading buffer. Equal amounts of eluted material were run in duplicate on 8.0% poly-acrylamide gels and western blotting was carried-out separately with anti-FLAG and anti-Ub antibodies. Ponceau S staining of both blots (only the ones for the anti-FLAG blots are presented) was performed to verify equivalent amount of recombinant proteins used in each binding reaction.
Microsome-based mobilization assay
Microsomes were prepared as described previously (Shcherbik et al., 2003) and placed in reactions containing 25 mM Tris HCl pH 7.5, 75 mM NaCl, 5 mM MgCl2, 3 mM ATP, 1 mM creatine phosphate, 0.05 units/μl creatine kinase, 20 μM DTT, 0.2 mM PMSF, and 0.5mM MG115 and 300 ng of recombinant proteins or complexes. For reactions containing Apyrase (Sigma), enzyme was added to a final concentration of 15 units/ml. Reactions were incubated at 27°C for 1hr and were separated into insoluble (membrane) and soluble fractions by ultracentrifugation at 50,000xg for 30 min at 4°C. The amount of Mga2p protein present in both fractions was determined by western blotting using anti-FLAG antibody. To assess ubiquitination status, proteins were extracted and/or immunopurified from the indicated fractions using anti-FLAG M2 coupled agarose (from both, soluble and microsomal fractions). Proteins were eluted in SDS-PAGE loading buffer, separated by SDS-PAGE, transferred on nitrocellulose membrane and probed with indicated antibodies.
Microsome-free in vitro segregation assay
Poly-Ubp120-p90 complexes of Mga2p and Spt23p were prepared as described above; except that FLAGMGA2HA and FLAGSPT23HA expression constructs harboring both amino-terminal FLAG and carboxy-terminal HA tags were used. After elution with FLAG peptide, complexes were immobilized on HA antibody-containing agarose (Sigma). Bead captured complexes were then placed in mobilization reaction buffer with the indicated recombinant proteins. Reactions were incubated at room temperature for 1 hr and pelleted by brief centrifugation. The amount of Mga2p/Spt23p in supernatant and on pelleted agarose beads was determined by western blotting.
Supplementary Material
Acknowledgments
This work is supported by NIH grant GM070769 to DSH. We thank S. Fuchs for providing a critical review of the manuscript. Also, we thank Hector Victoria for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auld KL, Brown CR, Casolari JM, Komili S, Silver PA. Genomic association of the proteasome demonstrate overlapping gene regulatory activity with transcription factor substrates. Mol Cell. 2006;21:861–871. doi: 10.1016/j.molcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Bays NW, Hampton RY. Cdc48-Ufd1-Npl4: stuck in the middle with Ub. Curr Biol. 2002;12:R366–371. doi: 10.1016/s0960-9822(02)00862-x. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Halawani D, Latterich M. p97: The Cell’s Molecular Purgatory? Mol Cell. 2006;22:713–717. doi: 10.1016/j.molcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Hitchcock AL, Krebber H, Frietze S, Lin A, Latterich M, Silver PA. The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol Biol Cell. 2001;12:3226–3241. doi: 10.1091/mbc.12.10.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Ishigaki S, Hishikawa N, Niwa J, Iemura S, Natsume T, Hori S, Kakizuka A, Tanaka K, Sobue G. Physical and functional interaction between Dorfin and Valosin-containing protein that are colocalized in ubiquitylated inclusions in neurodegenerative disorders. J Biol Chem. 2004;279:51376–51385. doi: 10.1074/jbc.M406683200. [DOI] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally JE, Chernova T, Wilkinson KD. Doa1 is a Cdc48 adapter that possesses a novel ubiquitin binding domain. Mol Cell Biol. 2006;26:822–830. doi: 10.1128/MCB.26.3.822-830.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Isaacson R, Kim HT, Silver PA, Wagner G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure. 2005;13:995–1005. doi: 10.1016/j.str.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Piwko W, Jentsch S. Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat Struct Mol Biol. 2006;13:691–697. doi: 10.1038/nsmb1122. [DOI] [PubMed] [Google Scholar]
- Pye VE, Dreveny I, Briggs LC, Sands C, Beuron F, Zhang X, Freemont PS. Going through the motions: the ATPase cycle of p97. J Struct Biol. 2006;156:12–28. doi: 10.1016/j.jsb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48UFD1/NPL4, a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. MolCell. 2006;21:261–269. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Zoladek T, Nickels JT, Haines DS. Rsp5p is required for ER bound Mga2p120 polyubiquitination and release of the processed/tethered transactivator Mga2p90. Curr Biol. 2003;13:1227–1233. doi: 10.1016/s0960-9822(03)00457-3. [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Kee Y, Lyon N, Huibregtse JM, Haines DS. A single PXY motif located within the carboxyl terminus of Spt23p and Mga2p mediates a physical and functional interaction with ubiquitin ligase Rsp5p. J Biol Chem. 2004;279:53892–53898. doi: 10.1074/jbc.M410325200. [DOI] [PubMed] [Google Scholar]
- Wang Q, Song C, Li CC. Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J Struct Biol. 2004;146:44–57. doi: 10.1016/j.jsb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Woodman PG. p97, a protein coping with multiple identities. JCell Sci. 2003;116:4283–4290. doi: 10.1242/jcs.00817. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y. Diverse function with a common regulator: ubiquitin takes command of an AAA ATPase. J Struct Biol. 2006;156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Adachi E, Fukiya K, Iwai K, Tanaka K. Glycoprotein-specific ubiquitin ligases recognize N-glycans in unfolded substrates. EMBO Rep. 2005;6:239–244. doi: 10.1038/sj.embor.7400351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


