Abstract
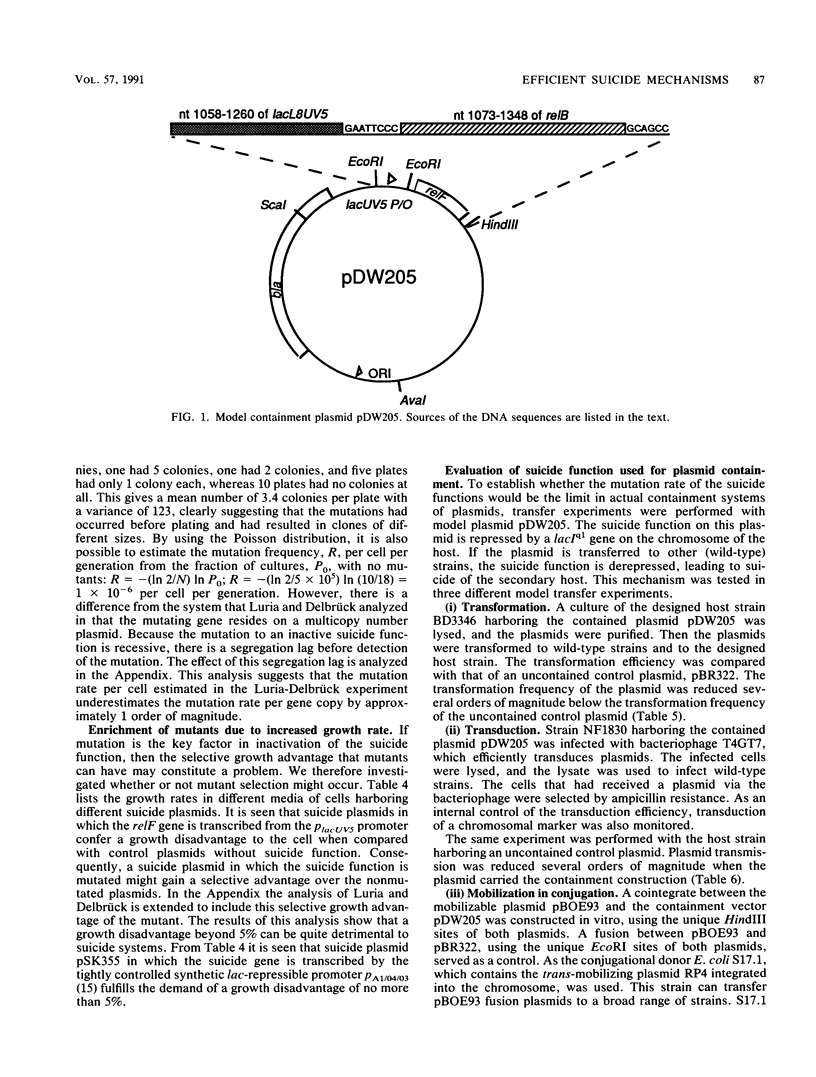
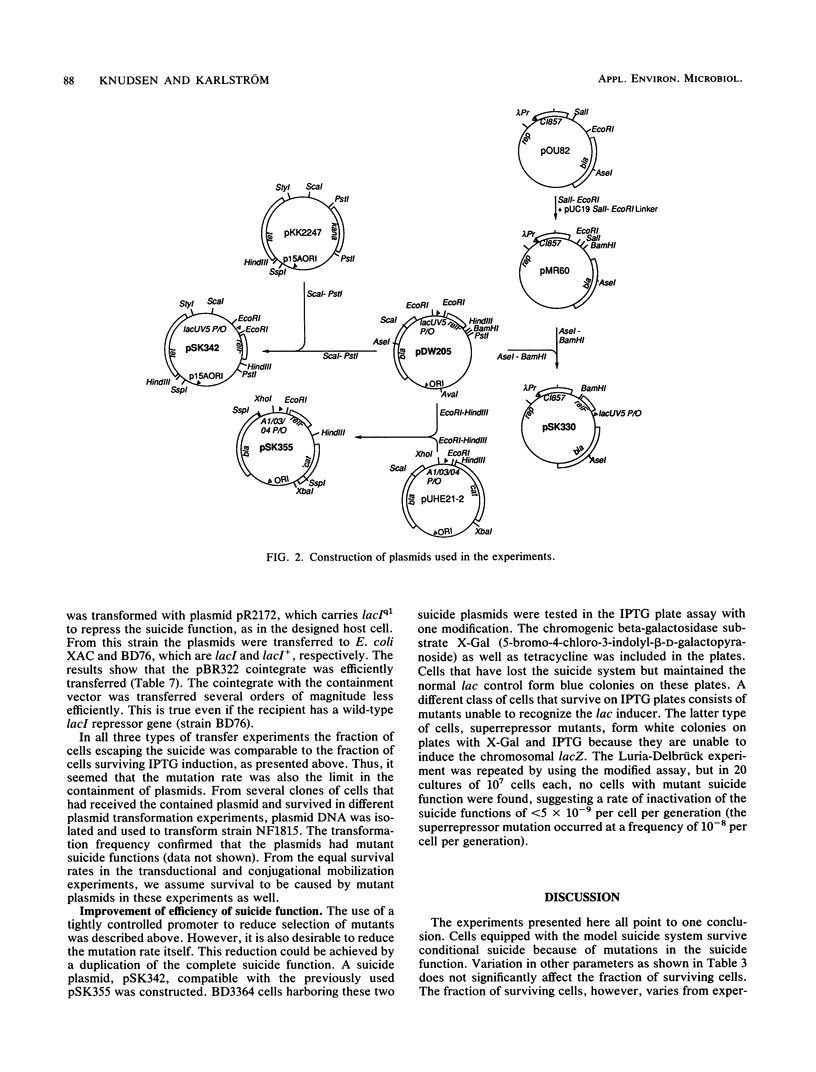
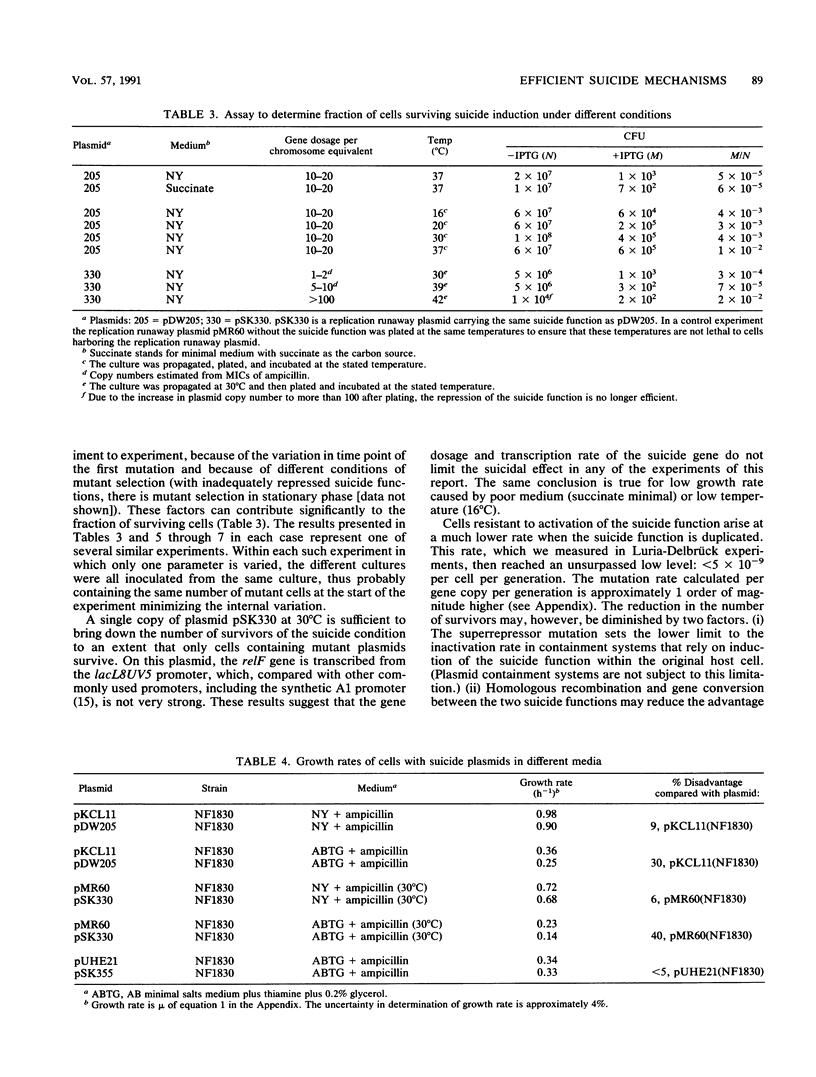
To optimize plasmid containment, we have systematically investigated the factors that limit the killing efficiency of a suicide system based on the relF gene from Escherichia coli controlled by inducible lac promoters and placed on plasmids. In induction experiments with this suicide system, killing efficiency was unaffected by temperature and growth medium; there was no requirement for great promoter strength or high plasmid copy number. We could demonstrate that the factors limiting killing were the mutation rate of the suicide function and the reduced growth rate caused by a basal level of expression of the suicide gene during normal growth, which can give a selective growth advantage to cells with mutated suicide functions. The capacity of the plasmid-carried killing system to contain the plasmid was tested in transformation, transduction, and conjugational mobilization. The rate of plasmid transfer detected in these experiments seemed too high to provide adequate biological containment. As expected from the induction experiments, plasmids that escaped containment in these transfer experiments turned out to be mutated in the suicide function. With lac-induced suicide as a test, the efficiency of the system was improved by tightening the repression of the suicide gene, thereby preventing selection of cells mutated in the killing function. Reduction of the mutational inactivation rate of the suicide system by duplication of the suicide function augmented the efficiency of the suicide dramatically. These results permit the construction of extremely efficient biological containment systems.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beckwith J., Grodzicker T., Arditti R. Evidence for two sites in the lac promoter region. J Mol Biol. 1972 Aug 14;69(1):155–160. doi: 10.1016/0022-2836(72)90031-9. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Bonekamp F., Clemmesen K., Karlström O., Jensen K. F. Mechanism of UTP-modulated attenuation at the pyrE gene of Escherichia coli: an example of operon polarity control through the coupling of translation to transcription. EMBO J. 1984 Dec 1;3(12):2857–2861. doi: 10.1002/j.1460-2075.1984.tb02220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAVALLI-SFORZA L. L. La sessualità nei batteri. Boll Ist Sieroter Milan. 1950 Sep-Oct;29(9-10):281–289. [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias L., Cervantes L., Covarrubias A., Soberón X., Vichido I., Blanco A., Kupersztoch-Portnoy Y. M., Bolivar F. Construction and characterization of new cloning vehicles. V. Mobilization and coding properties of pBR322 and several deletion derivatives including pBR327 and pBR328. Gene. 1981 Jan-Feb;13(1):25–35. doi: 10.1016/0378-1119(81)90040-8. [DOI] [PubMed] [Google Scholar]
- Davis B. D. Transcriptional bias: a non-Lamarckian mechanism for substrate-induced mutations. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5005–5009. doi: 10.1073/pnas.86.13.5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Bech F. W., Jørgensen S. T., Løbner-Olesen A., Rasmussen P. B., Atlung T., Boe L., Karlstrom O., Molin S., von Meyenburg K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986 Aug;5(8):2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Larsen J. E., Molin S. Stable inheritance of plasmid R1 requires two different loci. J Bacteriol. 1985 Jan;161(1):292–298. doi: 10.1128/jb.161.1.292-298.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzer M., Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
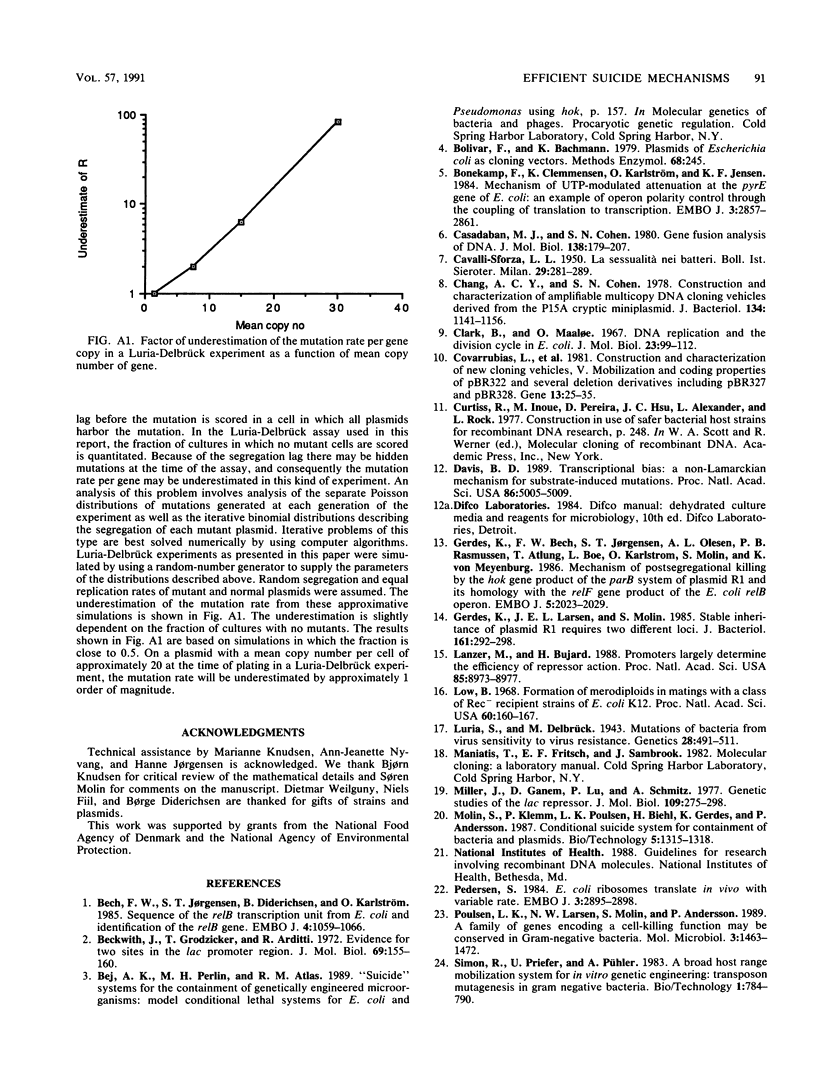
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Ganem D., Lu P., Schmitz A. Genetic studies of the lac repressor. I. Correlation of mutational sites with specific amino acid residues: construction of a colinear gene-protein map. J Mol Biol. 1977 Jan 15;109(2):275–298. doi: 10.1016/s0022-2836(77)80034-x. [DOI] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984 Dec 1;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen L. K., Larsen N. W., Molin S., Andersson P. A family of genes encoding a cell-killing function may be conserved in all gram-negative bacteria. Mol Microbiol. 1989 Nov;3(11):1463–1472. doi: 10.1111/j.1365-2958.1989.tb00131.x. [DOI] [PubMed] [Google Scholar]
- Stewart F. M., Gordon D. M., Levin B. R. Fluctuation analysis: the probability distribution of the number of mutants under different conditions. Genetics. 1990 Jan;124(1):175–185. doi: 10.1093/genetics/124.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Saito H. High-frequency transduction of pBR322 by cytosine-substituted T4 bacteriophage: evidence for encapsulation and transfer of head-to-tail plasmid concatemers. Plasmid. 1982 Jul;8(1):29–35. doi: 10.1016/0147-619x(82)90038-5. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Young K. Y., Edlin G. J., Konigsberg W. High-frequency generalised transduction by bacteriophage T4. Nature. 1979 Jul 5;280(5717):80–82. doi: 10.1038/280080a0. [DOI] [PubMed] [Google Scholar]
- von Meyenburg K., Jørgensen B. B., Nielsen J., Hansen F. G. Promoters of the atp operon coding for the membrane-bound ATP synthase of Escherichia coli mapped by Tn10 insertion mutations. Mol Gen Genet. 1982;188(2):240–248. doi: 10.1007/BF00332682. [DOI] [PubMed] [Google Scholar]


