Abstract
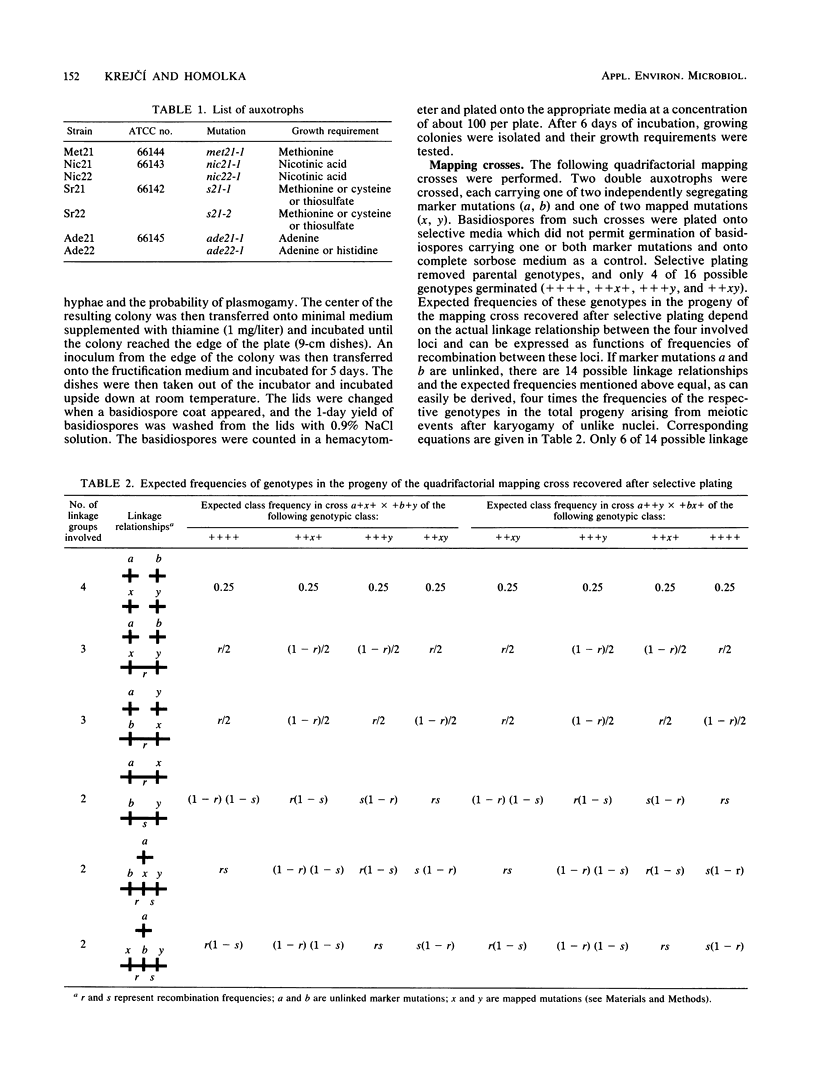
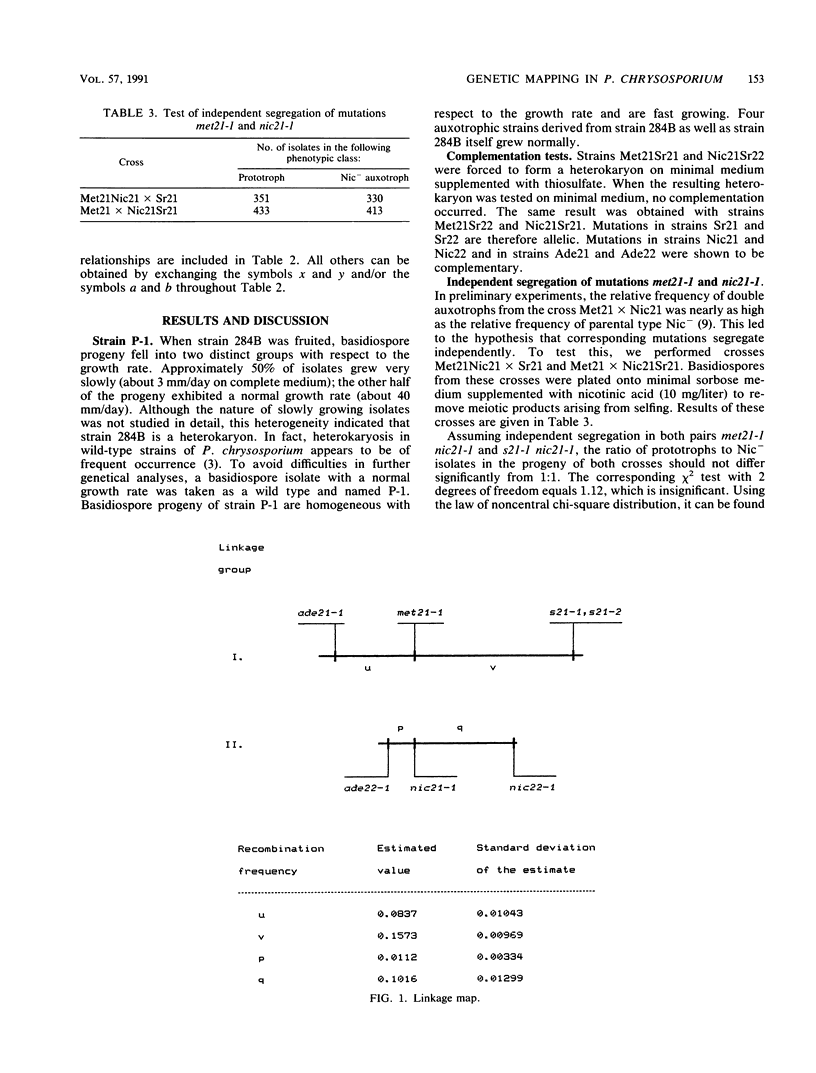
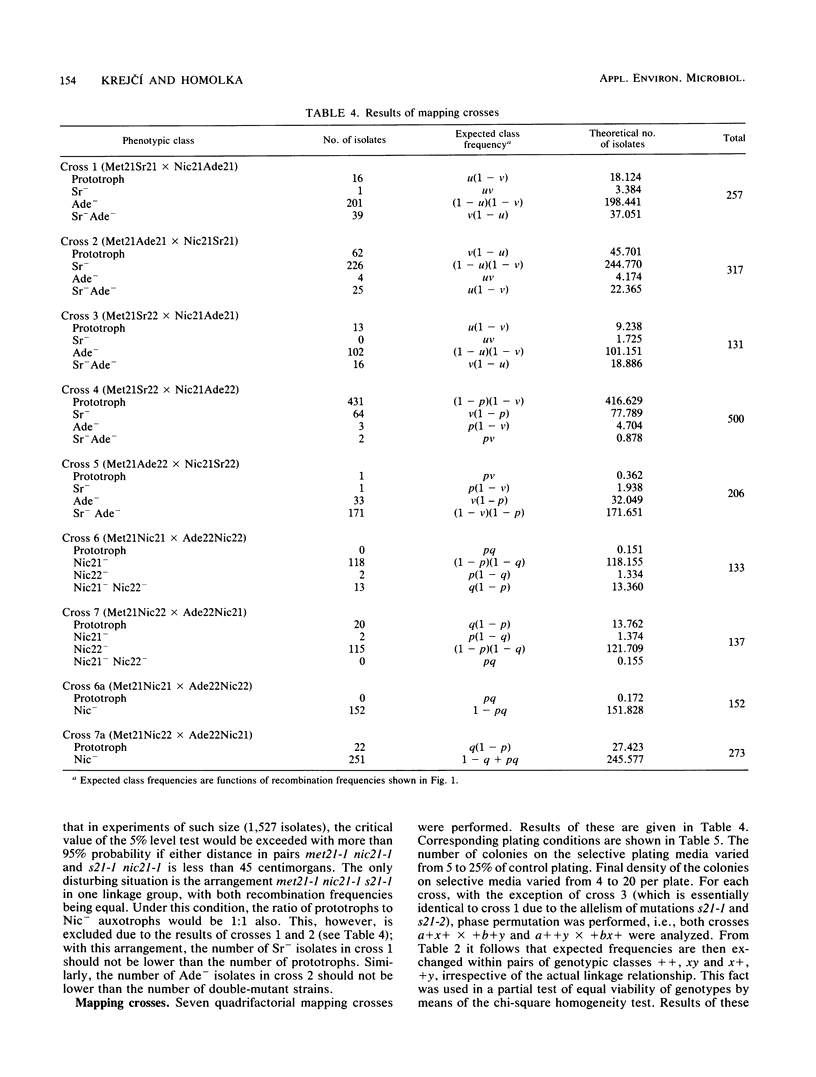
A method of meiotic segregation analysis based on recombinant selection in the homothallic basidiomycete Phanerochaete chrysosporium was developed. Using this method, we were able to reveal linkage relationships and to estimate recombination frequencies between seven mutations to auxotrophy. We detected two linkage groups, the first containing four and the second three of the seven mapped mutations.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alic M., Gold M. H. Genetic Recombination in the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1985 Jul;50(1):27–30. doi: 10.1128/aem.50.1.27-30.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic M., Kornegay J. R., Pribnow D., Gold M. H. Transformation by Complementation of an Adenine Auxotroph of the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1989 Feb;55(2):406–411. doi: 10.1128/aem.55.2.406-411.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic M., Letzring C., Gold M. H. Mating System and Basidiospore Formation in the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1987 Jul;53(7):1464–1469. doi: 10.1128/aem.53.7.1464-1469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M. H., Cheng T. M. Induction of colonial growth and replica plating of the white rot basidiomycete Phanaerochaete chrysosporium. Appl Environ Microbiol. 1978 Jun;35(6):1223–1225. doi: 10.1128/aem.35.6.1223-1225.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M. H., Cheng T. M., Mayfield M. B. Isolation and Complementation Studies of Auxotrophic Mutants of the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1982 Oct;44(4):996–1000. doi: 10.1128/aem.44.4.996-1000.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzbaur E. L., Andrawis A., Tien M. Structure and regulation of a lignin peroxidase gene from Phanerochaete chrysosporium. Biochem Biophys Res Commun. 1988 Sep 15;155(2):626–633. doi: 10.1016/s0006-291x(88)80541-2. [DOI] [PubMed] [Google Scholar]
- PONTECORVO G., ROPER J. A., HEMMONS L. M., MACDONALD K. D., BUFTON A. W. J. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Pribnow D., Mayfield M. B., Nipper V. J., Brown J. A., Gold M. H. Characterization of a cDNA encoding a manganese peroxidase, from the lignin-degrading basidiomycete Phanerochaete chrysosporium. J Biol Chem. 1989 Mar 25;264(9):5036–5040. [PubMed] [Google Scholar]
- Raeder U., Broda P. Meiotic segregation analysis of restriction site polymorphisms allows rapid genetic mapping. EMBO J. 1986 Jun;5(6):1125–1127. doi: 10.1002/j.1460-2075.1986.tb04336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeder U., Thompson W., Broda P. Genetic factors influencing lignin peroxidase activity in Phanerochaete chrysosporium ME446. Mol Microbiol. 1989 Jul;3(7):919–924. doi: 10.1111/j.1365-2958.1989.tb00241.x. [DOI] [PubMed] [Google Scholar]
- Raeder U., Thompson W., Broda P. RFLP-based genetic map of Phanerochaete chrysosporium ME446: lignin peroxidase genes occur in clusters. Mol Microbiol. 1989 Jul;3(7):911–918. doi: 10.1111/j.1365-2958.1989.tb00240.x. [DOI] [PubMed] [Google Scholar]
- Tien M., Tu C. P. Cloning and sequencing of a cDNA for a ligninase from Phanerochaete chrysosporium. Nature. 1987 Apr 2;326(6112):520–523. doi: 10.1038/326520a0. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Z., Zylstra G. J., Olsen R. H., Reddy C. A. Identification of cDNA clones for ligninase from Phanerochaete chrysosporium using synthetic oligonucleotide probes. Biochem Biophys Res Commun. 1986 Jun 13;137(2):649–656. doi: 10.1016/0006-291x(86)91127-7. [DOI] [PubMed] [Google Scholar]



