Abstract
We have identified a sine oculis gene in the planarian Girardia tigrina (Platyhelminthes; Turbellaria; Tricladida). The planarian sine oculis gene (Gtso) encodes a protein with a sine oculis (Six) domain and a homeodomain that shares significant sequence similarity with so proteins assigned to the Six-2 gene family. Gtso is expressed as a single transcript in both regenerating and fully developed eyes. Whole-mount in situ hybridization studies show exclusive expression in photoreceptor cells. Loss of function of Gtso by RNA interference during planarian regeneration inhibits eye regeneration completely. Gtso is also essential for maintenance of the differentiated state of photoreceptor cells. These results, combined with the previously demonstrated expression of Pax-6 in planarian eyes, suggest that the same basic gene regulatory circuit required for eye development in Drosophila and mouse is used in the prototypic eye spots of platyhelminthes and, therefore, is truly conserved during evolution.
Keywords: homeobox, eye morphogenesis, platyhelmint, eye evolution
The study of the genetic network that regulates the development of the Drosophila visual system has resulted in the identification of several transcription factors and other nuclear proteins that are required for the specification of early eye morphogenesis (1–4). These factors seem to act in a hierarchy in which sine oculis (so) is regulated directly by Pax-6 (5, 6), the master control function. In turn, so requires eyes absent (eya), encoding a nuclear protein (7), to induce ectopic eyes (4). This genetic pathway has been established in Drosophila (8), but homologous proteins also regulate eye development in vertebrates, suggesting that this regulatory network is old, is conserved in evolution, and has been adapted to the control of development of different visual systems found in both clades (9). Both the identification and functional characterization of homologous genes in more primitive organisms, such as the platyhelminthes, will help to clarify the age and extent of conservation of this genetic cascade.
Sine oculis is a homeobox-containing gene that is required for the development of the visual system in Drosophila (10, 11). A murine homologue, Six3, is expressed in the developing eye (12). In both of these model systems, so and Six are expressed early in eye development as well as in other structures. Combined overexpression of so and eya in Drosophila induces ectopic eyes (4), whereas, in vertebrates, Six3 overexpression results in ectopic lens formation (13, 14). Planarians (Platyhelminthes; Turbellaria; Tricladida) are located at the base of the Lophotrochozoa Protostomia clade (15, 16). The eye spots of planarians are one of the most ancestral and simple types of visual systems, close to the prototypic eye proposed by Charles Darwin (see ref. 8). The planarian eye spots consist of two cell types: a bipolar nerve cell with a rhabdomere as a photoreceptive structure and a cup-shaped structure composed of pigment cells (17). During head regeneration, new eye spots are formed from precursor cells that differentiate into both cell types in a restricted area of the newly regenerated tissue or blastema. Previous studies of this regenerative process show a clear expression of planarian Pax-6 (GtPax-6) in both visual cell types (18).
In the current study, we address the hypothesis that a Pax-6-regulated network is conserved in evolution, and as a consequence, Girardia tigrina eye development requires a sine oculis homologue. We report the identification of an so gene from the planarian G. tigrina (Gtso). The high degree of amino acid sequence identity in the sine oculis domain and in the homeodomain suggests that Gtso is orthologous to known invertebrate so genes and belongs to the Six2 gene family. The expression of Gtso in intact and regenerating planarians suggests a putative role in development and maintenance of the eye. RNA interference (RNAi) experiments provide functional evidence that Gtso is essential for maintenance of the differentiated state of photoreceptor cells and for eye regeneration. These findings suggest that the basic elements of the genetic pathway are conserved in these prototypic eyes.
Materials and Methods
Species.
The planarians used in this study belong to an asexual race (class A; ref. 19) of the species G. tigrina. Specimens were collected near Barcelona. They were maintained in spring water. Organisms starved for 2 weeks were used in all experiments. Planarians 9- to 10-mm-long were cut prepharyngeally according to the method described in ref. 20 and were left regenerating in Petri dishes with spring water in the dark at 17°C.
Isolation of the Gtso Gene.
An so fragment was amplified by PCR from planarian genomic DNA with a pair of degenerate primers corresponding to amino acids conserved between Six1, Six2, Six3, and so. The sense primer (so1), consisting of a degenerate sequence corresponding to amino acid sequence WDGEET with 5′ clamp sequences and an XhoI site [gta ctc gag tgg ga(t,c) gg(a,c,g,t) ga(a,g) ga(a,g) ac], was used. The antisense primer (so2) used consisted of a degenerate sequence corresponding to amino acid sequence QRDRAA with 5′ clamp sequences and an XbaI site [ccg tct aga c(a,c,g,t)g cic (g,t)(a,g)t cic (g,t)(t,c)tg]. PCRs were performed in 100-μl volumes in the presence of 0.5 μg of genomic DNA. The cycling program consisted of 5 cycles (94°C for 1 min, 46°C for 2 min, and 72°C for 3 min with ramping times of 1 min to 94°C, 1 min to 46°C, and 2 min 30 s to 72°C) and 30 cycles (94°C for 1 min, 65°C for 1 min, and 72°C for 1 min). The identity of the Gtso fragment was confirmed by sequencing. Based on this sequence, Gtso-specific nested primers were designed for the amplification of the full-length Gtso by rapid amplification of cDNA ends by PCR with the Marathon kit (CLONTECH). Total RNA from head- and tail-regenerating animals was isolated as described in ref. 21. Poly(A)+ RNA was isolated with the Oligotex mRNA Microkit (Qiagen, Chatsworth, CA) following the manufacturer's instructions. Amplified fragments were cloned in the TA cloning vector pCR2.1 (Invitrogen). Both strands of the cDNA were sequenced twice in their entirety by dideoxy sequencing and primer walking with the ABI PRISM kit (Perkin–Elmer) and with the Sequenase 2 kit (United States Biochemical).
Phylogenetic Analysis of the Gtso Sine Oculis Homeodomains.
The phylogenetic trees of sine oculis homeodomain sequences were inferred by using the clustalx package. Sequences were aligned with the software clustalx, and refined alignment was done manually. The Kimura's equation was used for the evolutionary distances (22), and the neighbor-joining method was used for the tree construction. Sequences were obtained from the EMBL GenBank and the DNA Data Bank of Japan.
Northern Blotting and Whole-Mount in Situ Hybridization.
Northern blot analyses were performed by standard procedures (23). Whole-mount in situ hybridizations were carried out with intact planarians and at different regenerative stages according to the method described in ref. 24. Fixed and bleached planarians were treated with proteinase K (20 μg/ml) for different times (between 8 to 15 min) depending on their size and regenerative stage. Hybridizations were carried out at 55°C for 60 h. After color development, the samples were postfixed in 4% (vol/vol) paraformaldehyde/PBS, cryoprotected in sucrose solutions, embedded in Tissue Freezing Medium (Jung, Leica, Nussloch, Germany), and sectioned with a cryomicrotome.
Synthesis and Microinjection of Double-Strand RNA (dsRNA).
dsRNA was synthesized as described in ref. 25. The opsin clone op-250 (GenBank accession no. AJ251660) was digested with XhoI or BamHI to synthesize antisense (T3) or sense (T7) RNAs. Gtso clones so-5′ and so-3′-2 (GenBank accession no. AJ251661) were digested with HindIII or XbaI to synthesize antisense (T3) or sense (T7) RNAs. Planarians were injected with 1010 molecules of dsRNA or water with a Drummond Scientific (Broomall, PA) Nanoject injector. All of the injected planarians were kept at 17°C. Adult planarians were injected into the parenchyma close to the eye region, and 3-day head-regenerating pieces were injected into the postblastema tissue, the old tissue close to the wound. The volumes of injection were 23 nl each. At different stages of regeneration, the injected fragments were photographed and fixed, and whole-mount in situ hybridizations for Gtso or opsin were performed.
Results
Isolation and Sequence Comparison of Gtso.
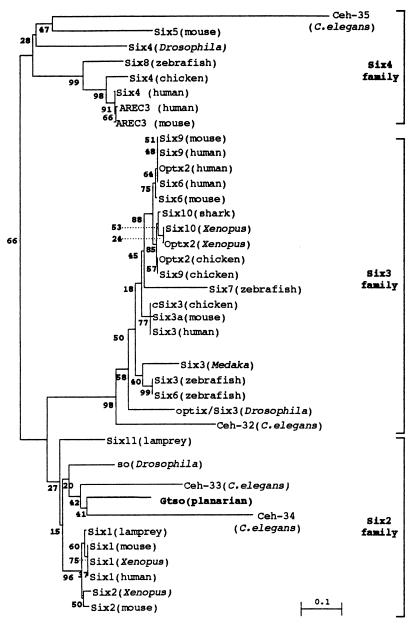
Initial isolation and partial characterization of a planarian so homologue was achieved by PCR amplification of genomic DNA with a pair of degenerate primers. A complete cDNA of 1,522 bp was identified by nested rapid amplification of both cDNA ends by PCR. The ORF encodes 435 amino acids with two regions of high sequence conservation in the deduced protein: the sine oculis domain and the homeodomain. Comparison with so homeodomain sequences of other species (Fig. 1) shows the highest sequence identity to the so/Six-2 family proteins, which share the consensus tetrapeptide (ETSY; ref. 28) and other residues scattered through the homeodomain. The sequence conservation is not only restricted to the homeodomain but also includes approximately 117 amino acids of the 5′ flanking region (the sine oculis domain). This domain is less conserved than the homeodomain, but its comparative amino acid analysis can group Gtso into the so/Six-2 family.
Figure 1.
Alignment of so/Six homologues homeodomain amino acid sequences of mouse used to represent vertebrates and of Drosophila and Caenorhabditis elegans used to represent invertebrates. The secondary structure of the domains is shown at the top of the figure. Amino acid identities for representative genes of the so/Six family are compared with the Drosophila so homeodomain (10). Sequences used in this comparison include: mouse Six1, Six2, and Six3 (12); Six4 (26); Drosophila optix/Six3 and Six4 (27, 28); and the nematode C. elegans Ceh-32, Ceh-33, and Ceh-34 (EMBL database). The tetrapeptides used to classify the sine oculis homeodomains in three families are indicated in bold and boxed. Percentages of sequence identity (%ID) and similarity (%S) were determined by comparison to Drosophila so and are indicated to the right of the sequences.
The C-terminal region comprises 229 amino acids rich in serine (14%), asparagine (13.5%), proline (7%), and threonine (8%), suggesting the presence of transactivating functions (29), and 30% of these serines, and other less frequent residues scattered throughout the sequence, are also present in the same position of the different so homologous proteins. Another feature of this protein is the presence of several amino acid doublets and some repeats of tetrapeptides and pentapeptides, for which the significance is not known.
Phylogenetic trees were constructed for sine oculis homeodomain sequences. The distances were computed with Kimura's equation (22). We can observe the clustering of Six genes in three main groups or families where the Gtso is grouped with members of the so/Six-2 gene family (Fig. 2).
Figure 2.
Phylogenetic unrooted tree of sine oculis homeodomains. Bootstrap values of 1,000 runs are indicated as percentages at the nodes. The planarian Gtso homeodomain clearly clusters with the other sine oculis genes from the so/Six2 family. Scale bar, genetic distance.
Gtso Expression in Intact and Regenerating Adults.
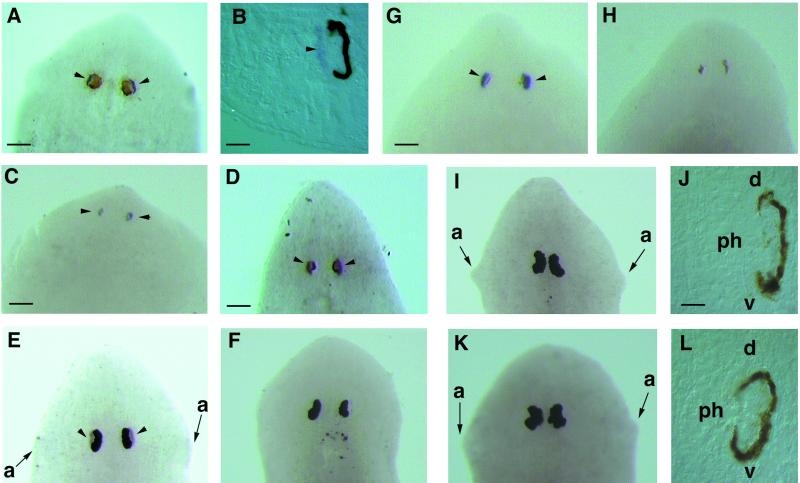
Gtso expression in intact and regenerating planarians was analyzed by Northern blotting and in situ hybridization. Northern blot analysis shows a unique transcript of around 1.5 kilobases in adults and in regenerative stages without any evidence for differential splicing (results not shown). The Gtso spatial expression was determined by whole-mount in situ hybridization and by posterior transversal cryosectioning of the same intact adults and regenerating planarians after hybridization. In adults, Gtso was expressed continuously and uniformly in the photoreceptor cell bodies, whereas the rhabdomeric region of the photoreceptor cells was negative. No signal was observed in the pigment cells (Fig. 3 A and B). During the early stages of head regeneration, Gtso expression was detected in a group of photoreceptor cells close to the dorsal epidermis (Fig. 3C), which constitutes the earliest visible sign of eye regeneration. This early expression in the eye primordia was maintained throughout regeneration (Fig. 3D).
Figure 3.
(A–D) Dorsal view of Gtso expression by whole-mount in situ hybridization. Intact head (A) and cryosection (B) of the same head, as well as head-regenerating planarian adults at 7 days (C) and 14 days (D). Arrowheads indicate the localization of Gtso mRNA indicated by blue signal in the photoreceptor cells. Close to these, it can be observed that the brown pigmented cells are located more centrally. After 7 days of regeneration, when the first pigmented cells appear, a clear blue signal of Gtso expression can be observed. This signal is maintained throughout the whole regenerative process. (E–H) Opsin mRNA inhibition in photoreceptors by G. tigrina opsin dsRNA injection: this control organism shows the opsin mRNA distribution in the photoreceptor cells (arrowheads) (E); this adult organism, 24 h after injection, does not show any accumulation of endogenous opsin mRNA (F); control planarians after 7 days of head regenerating express opsin strongly (G); and opsin dsRNA-injected organisms in the same regenerative stage express no opsin (H). (I–L) Adult heads injected with Gtso dsRNA. (I and J) Whole-mount in situ hybridization and cryosections with Gtso riboprobes of organisms injected 24 h previously; no expression of Gtso can be observed. (K and L) Whole-mount in situ hybridization and cryosections with opsin riboprobes of organisms injected 7 days previously; no expression of opsin can be observed in the remaining photoreceptor cells. a, auricle; d, dorsal; ph, photoreceptor cells; v, ventral. [Bars = 300 μm (D), 200 μm (A, E, F, I, and K), 150 μm (C, G, and H), 40 μm (B), and 20 μm (J and L).]
RNAi Disrupts Endogenous Gene Expression in Intact Adult and Regenerating G. tigrina Tissues.
To test the efficacy of RNAi in planarian G. tigrina, we injected heads of intact animals and regenerating pieces with approximately 1010 copies of 250-bp opsin dsRNA synthesized from a G. tigrina opsin cDNA clone. As described for Schmidtea mediterranea (25), 24 h after injection, no opsin mRNA was observed by whole-mount in situ hybridization in the adult differentiated photoreceptor cells. This inhibitory effect lasts for up to 3 weeks, whereas the controls maintain a continuous and specific expression (Fig. 3 E and F). Water-injected controls start to differentiate eyes after 7 days at 17°C, but the opsin dsRNA-injected specimen did not show any sign of opsin expression (Fig. 3 G and H). This result shows that the dsRNA injected into the parenchymal cavity of G. tigrina quickly leads to the absence of the mRNA of a gene highly expressed in differentiated photoreceptor cells.
Loss of Function of Gtso by RNAi Injection Produces a No-Eye Phenotype.
Because Gtso is expressed specifically in the photoreceptor cells, we tried to determine whether the introduction of dsRNA into head-regenerating planarians could reduce endogenous expression levels of Gtso and alter the formation of the eye structures. To observe the effects exerted by the Gtso dsRNA injections, nonregenerating and regenerating adult organisms were analyzed at different times and compared with the water-injected controls. Injection of Gtso dsRNA molecules into the adult differentiated eye did not cause any detectable change in the pigment cells, whereas the photoreceptor cells also remained intact but lost Gtso expression 24 h after injection (Fig. 3 I and J). A second effect observed in the injected organisms was the loss of opsin expression (Fig. 3 K and L), indicating that Gtso is required for the maintenance of the differentiated state of the photoreceptor cells. The control organisms maintained Gtso and opsin expression, which is indicative of the presence of intact and functional photoreceptor cells (not shown).
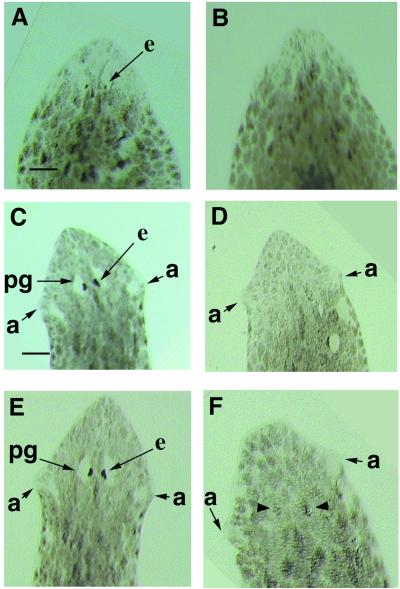
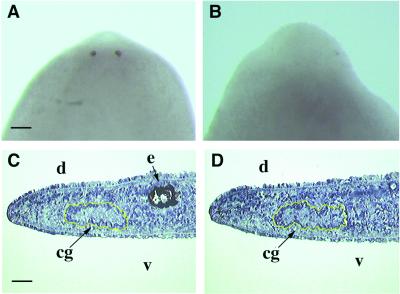
Distinct eye structures can be observed after 7 days in the regenerating controls. Pigment cells are detected by bright field microscopy (Fig. 4A), and differentiated photoreceptor cells are identified by whole-mount in situ hybridization with the opsin riboprobe (Fig. 5A). Gtso dsRNA-injected regenerating planarians had the same size and differentiation level of the blastema as the controls, with well differentiated auricles on the either side of the head. However, the eyes did not differentiate even after 3 weeks of regeneration after a single injection (Fig. 4 B and D). In the injected animals, neither pigment cells nor photoreceptors form (Fig. 5B), whereas the 7- and 14-day regenerating controls show completely regenerated eyes with their periglobular nonpigmented area in the dorsal epidermis above the eyes (Fig. 4 A and C). All of the 30 Gtso dsRNA-injected regenerating planarians in three independent experiments differentiated their dorsal blastema without any type of periglobular nonpigmented area, thus indicating that this structure may be induced by the differentiated eyes (Fig. 4 B, D, and F). The inhibition of eye regeneration is very consistent. All of the 30 head-regenerating fragments had a no-eye phenotype, even 3 weeks after injection. The continued maintenance of such eye inhibition in the head blastemas required a reinjection treatment every 3 weeks, because, after 4 or 5 weeks without injection, new eyes start to regenerate in the already differentiated heads of some organisms (Fig. 4F). Therefore, the inhibition is transient. To exclude any indirect effect on eye formation caused by the loss of the Gtso function by inhibiting the differentiation of the cephalic nervous system, the regenerated cephalic ganglia in the Gtso dsRNA-injected organisms were analyzed histologically and compared with the controls. No differences in morphology and size of the cephalic ganglia were detected (Fig. 5 C and D).
Figure 4.
Inhibition of eye-regenerative capacity by Gtso dsRNA injection into the regenerative postblastemas. All of the organisms are in dorsal view. (A, C, and E) Control organisms at the same regenerative stage as those injected (B, D, and F). Bright-field images showing the eye differentiation stages with the formation of a periglobular unpigmented area completely absent in the injected organisms: after 7 days of regeneration (A and B); after 14 days of regeneration (C and D); and after 28 days of regeneration (E and F). (B and D) No eye or periglobular unpigmented area can be observed in the injected organisms. (F) Heads injected 28 days earlier started to differentiate the eyes (arrowheads). a, auricle; e, eye spot; pg, periglobular unpigmented area. [Bars = 400 μm (C, D, and E) and 200 μm (A, B, and F)].
Figure 5.
Gtso dsRNA injection in the regenerative postblastemas inhibits photoreceptor and pigment eye cells but does not affect regeneration of cephalic ganglia. Control (A) and injected (B) organisms. Whole-mount in situ hybridization with opsin riboprobes was performed to visualize the early inhibitory effect of Gtso dsRNA in the photoreceptor cell differentiation after 7 days of regeneration. Control (C) and injected (D) organisms. Sagittal cryosections of differentiated head blastemas after 14 days, stained with toluidine blue, show the same type of cephalic ganglia differentiation, encircled in yellow. cg, cephalic ganglia; d, dorsal; e, eye spot; v, ventral. [Bars = 150 μm (A and B) and 100 μm (C and D)].
Discussion
The planarian G. tigrina has a bona fide so/Six-2 gene. The phylogenetic analysis shows clustering of Gtso with the other so/Six2 genes at a very high probability. This notion is corroborated further by the conservation of specific residues in the homeodomain, allowing us to consider it as an orthologue of the so/Six-2 family. The sequence identity is essentially confined to both the homeodomain, in which most of the so/Six-2 specific residues are conserved, and to the N-terminally located sine oculis domain. In the C-terminal region, several series of amino acid-rich regions related to transactivation domains are also found. The recent isolation of a planarian gene orthologous to the Six-3 family (D.P., J.G., and E.S., unpublished work) indicates the presence of at least two families of sine oculis proteins in the Lophotrochozoa group.
The second question addressed in this study deals with the role of sine oculis in eye development. In planarians, the sine oculis gene Gtso is continuously expressed in adult eyes. Initial expression also coincides with the first signs of eye differentiation during cephalic regeneration. Similar observations have been described for the planarian Pax-6 gene DtPax-6 (18). The expression of developmental regulatory genes in adults is usual, because planarians show great morphological plasticity in the continuous growth and regression or regeneration processes. The same role can be observed for DtPax-6 and Gtso, which are important regulators in eye development and regeneration.
Because heads that fail to regenerate eyes after Gtso dsRNA injection contain normal differentiated cephalic ganglia and auricles, we can assume that this loss of function has an effect exclusively on the process of eye formation. The maintenance of such eye inhibition in the head blastemas requires dsRNA reinjection at 3-week intervals, thus indicating that the head is always competent for eye regeneration. Gtso is expressed in the photoreceptors of the prototypic differentiated eyes of G. tigrina. Furthermore, RNAi induced loss of function indicates a crucial function of Gtso in early eye determination. These two observations provide further support for a dual role of so/Six genes in eye development, namely in early determination and in neuronal differentiation according to the Drosophila model (4). The fact that the adult differentiated eye of G. tigrina shows no morphological defects induced by RNAi may be caused by a slow turnover of the eye cells. The loss of opsin expression in the photoreceptor cells can be interpreted in several ways. According to the model proposed in refs. 3 and 4 in which early eye development in Drosophila is regulated by a network of interacting genes, including sine oculis, it is conceivable that the loss of Gtso by RNAi results in a disruption of this network and indirectly to the loss of opsin expression. Alternatively, because sine oculis is also expressed in the differentiated photoreceptors of Drosophila, it is possible that the sine oculis genes of both Drosophila and Girardia directly regulate the opsin expression, and as a consequence, the loss of Gtso leads to the loss of opsin.
The coexpression of DtPax-6 and Gtso at the same regeneration stages in the same precursor visual cells and in the differentiated photoreceptor cells, in addition to the essential and specific function of Gtso during eye regeneration, provides additional support for the evolutionary conservation of the initial genetic pathway in eye determination of triploblastic metazoans.
Acknowledgments
We thank K. Agata for teaching us the whole-mount in situ hybridization method, A. Sánchez and P. Newmark for providing the S. mediterranea opsin partial sequence, and M. Marsal and C. Gonzalez for helping with the whole-mount in situ hybridization. This work was supported by grants from the Swiss National Science Foundation and the Kantons of Basel (to W.J.G.), by grants from the Dirección General de Investigación Cieutilìca y Técuica (to E.S.) (Ministerio de Educación y Ciencia, Spain, PB95-0579 and PB98-1261-C02-01), and by a Formación de Personal Investigador fellowship (to D.P.) from Universitat de Barcelona.
Abbreviations
- dsRNA
double-strand RNA
- RNAi
RNA interference
Footnotes
References
- 1.Quiring R, Walldorf U, Kloter U, Gehring W J. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 2.Halder G, Callaerts P, Gehring W J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 3.Shen W, Mardon G. Development (Cambridge, UK) 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- 4.Pignoni F, Hu B, Zavitz K H, Xiao J, Garrity P A, Zipursky S L. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- 5.Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring W J. Development (Cambridge, UK) 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- 6.Niimi T, Seimiya M, Kloter U, Flister S, Gehring W J. Development (Cambridge, UK) 1999;126:2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- 7.Bonini N M, Leiserson W M, Benzer S. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- 8.Gehring W J, Ikeo K. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- 9.Treissman J E. BioEssays. 1999;21:843–850. doi: 10.1002/(SICI)1521-1878(199910)21:10<843::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 10.Cheyette B. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 11.Serikaku M A, O'Tousa J E. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver G, Mailhos A, Wehr R, Copeland N G, Jenkins N A, Gruss P. Development (Cambridge, UK) 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 13.Oliver G, Loosli F, Köster R, Wittbrodt J, Gruss P. Mech Dev. 1996;60:233–239. doi: 10.1016/s0925-4773(96)00632-6. [DOI] [PubMed] [Google Scholar]
- 14.Loosli F, Köster R W, Carl M, Krone A, Wittbrodt J. Mech Dev. 1998;74:159–164. doi: 10.1016/s0925-4773(98)00055-0. [DOI] [PubMed] [Google Scholar]
- 15.Carranza S, Baguñà J, Riutort M. Mol Biol Evol. 1997;14:485–497. doi: 10.1093/oxfordjournals.molbev.a025785. [DOI] [PubMed] [Google Scholar]
- 16.Bayascas J R, Castillo E, Saló E. Dev Genes Evol. 1998;208:467–473. doi: 10.1007/s004270050204. [DOI] [PubMed] [Google Scholar]
- 17.Kishida Y. Sci Rep Kanazawa Univ. 1967;12:75–110. [Google Scholar]
- 18.Callaerts P, Muñoz-Marmol A M, Glardon S, Castillo E, Sun H, Li W H, Gehring W J, Saló E. Proc Natl Acad Sci USA. 1999;96:558–563. doi: 10.1073/pnas.96.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribas M, Riutort M, Baguñà J. J Zool. 1989;218:609–626. [Google Scholar]
- 20.Saló E, Baguñà J. J Embryol Exp Morphol. 1984;83:63–80. [PubMed] [Google Scholar]
- 21.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 22.Nei M, Koehn R. In: The Neutral Theory of Molecular Evolution. Nei M, Koehn R, editors. Sunderland, MA: Sinauer; 1983. pp. 208–233. [Google Scholar]
- 23.Garcia-Fernàndez J, Baguñà J, Saló E. Proc Natl Acad Sci USA. 1991;88:7338–7342. doi: 10.1073/pnas.88.16.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umesono Y, Watanabe K, Agata K. Dev Growth Differ. 1997;39:723–727. doi: 10.1046/j.1440-169x.1997.t01-5-00008.x. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez A, Newmark P A. Proc Natl Acad Sci USA. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawakami K, Ohto H, Ikeda K, Roeder R G. Nucleic Acids Res. 1996;24:303–310. doi: 10.1093/nar/24.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toy J, Yang J M, Leppert G S, Sundin O H. Proc Natl Acad Sci USA. 1998;95:10643–10648. doi: 10.1073/pnas.95.18.10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo H, Curtiss J, Mlodzik M, Fjose A. Mech Dev. 1999;83:127–139. doi: 10.1016/s0925-4773(99)00045-3. [DOI] [PubMed] [Google Scholar]
- 29.Ptashne M. Nature (London) 1998;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]