Abstract
We report an extreme morphological difference between Drosophila sechellia and related species of the pattern of hairs on first-instar larvae. On the dorsum of most species, the posterior region of the anterior compartment of most segments is covered by a carpet of fine hairs. In D. sechellia, these hairs have been lost and replaced with naked cuticle. Genetic mapping experiments and interspecific complementation tests indicate that this difference is caused, in its entirety, by evolution at the ovo/shaven-baby locus. The pattern of expression of the ovo/shaven-baby transcript is correlated with this morphological change. The altered dorsal cuticle pattern is probably caused by evolution of the cis-regulatory region of ovo/shaven-baby in the D. sechellia lineage.
Most recent studies of evolutionary developmental biology have focused on establishing correlations between macroevolutionary changes in morphology and alterations in gene expression patterns (1–4). Although such studies suggest possible modes of developmental change underlying phenotypic evolution (and have additional uses in phylogeny reconstruction and determination of homology), distant taxonomic comparisons provide limited insight into how development evolves in natural populations. In particular, such comparisons do not identify the individual mutations altering developmental processes that were initially exposed to natural selection. This link is required to connect population processes to evolutionary patterns and is best addressed by determining the mutations causing phenotypic evolution within and between closely related species.
Classic evolutionary theory predicts that interspecific differences result from the accumulation of multiple mutations, each of small effect (5), although more recent theoretical developments have argued for a distribution of effects at a smaller number of loci accounting for most variation (6). Most observations of genetic differences between Drosophila species are broadly consistent with this latter expectation. For example, evolution at multiple loci underlies differences in the shape of the male genital arch (7, 8), differences in the acoustic mating signal (9), and the causes of male sterility (10, 11) between Drosophila simulans and Drosophila mauritiana. Likewise, D. sechellia's resistance to the toxic morinda fruit has a polygenic basis (12). Divergence in male secondary sexual traits between the Hawaiian drosophilids, Drosophila heteroneura and Drosophila silvestris, is also caused by evolution at multiple loci (13). In contrast, the intraspecific sitter/rover behavioral polymorphism of Drosophila melanogaster is caused by variation at a single locus (14). In addition, a surprising number of traits in populations exposed to recent strong artificial selection have provided evidence for evolution through changes in one or few genes of large effect [e.g., insecticide resistance (15) and maize evolution (16)]. Despite these examples of large single gene effects, it remains unclear how often interspecific differences are generated by the evolution of one or few loci.
Inspired by the discovery of Dickinson and coworkers (17) that the pattern of hairs on the dorsum of the first-instar larva varies dramatically between species of the Drosophila virilis group, we examined larvae from species of the D. melanogaster species subgroup for variation in hair patterning. Dickinson et al. found that in most species of the D. virilis group, most body segments possess three rows of robust denticles and a large lawn of fine hairs, a pattern similar to that found in D. melanogaster. Four species, however, produce only the robust denticle belts, with naked cuticle replacing the lawn of fine hairs. We have discovered that D. sechellia displays a similar phenotype, with each segment containing several rows of robust denticles, and a large region of naked cuticle. All other species of the D. melanogaster species group possess a lawn of fine hairs instead of naked cuticle. Here we document this variation and present genetic evidence that this difference between D. sechellia and its close relatives is caused in its entirety by evolution at the ovo/shaven-baby (ovo/svb) locus.
Materials and Methods
Fly Stocks.
Flies were maintained at 25°C on standard cornmeal agar. Wild-type lines of D. mauritiana (0241.5 and 0241.6) and D. sechellia (0248.2, 0248.3, 0248.4, 0248.5, and 0248.15) were obtained from the Species Stock Center (Bowling Green, OH). Stocks of D. melanogaster (Oregon-R) and D. simulans (Tsimbazaza and y1w1f2) were obtained from the Ashburner lab (University of Cambridge). The stock w1svb1/FM7 was provided by the Nüsslein–Volhard lab (Tübingen, Germany). We generated the recombinant y1w1svb1. Deficiency kit DK1, a collection of 41 deficiencies covering most of the X chromosome, as well as svb2/FM7, Df(1)cho2, Df(1)RC40, Df(1)HC244, Df(1)bi-DL1, Df(1)bi-D2, and Df(1)bi-DL2, were obtained from the Bloomington Stock Center.
Cuticle Preparations and Microscopy.
First-instar larvae were mounted as described (18) for microscopic examination of cuticle phenotypes. Dorsal hair patterns were imaged from standard cuticle preparations on a confocal microscope (Leica SP) (larval cuticles autofluoresce when exposed to the Argon laser).
Crosses.
Interspecific crosses were performed after maintaining virgin females with females of the opposite species for several days (19). Females of the second species, whose wings had been clipped for identification, were removed before adding males. Eggs or larvae were collected from apple juice plates (18). D. sechellia females laid eggs on normal medium supplemented with octanoic acid (Sigma) (240 μl per 100 g) (20).
PCR Cloning.
Genomic DNA was prepared by standard techniques (21). PCR primers were designed for the D. melanogaster ovo/svb second exon (22): 5′-ATTGCCTCCGTTTTATGAGA and 5′-TGCTCCAGTAAATGATCGGT. PCR was carried out with standard reagents (Boehringer–Mannheim), and the amplification products were cloned by using the pGEM-T Easy kit (Promega).
In Situ Hybridization.
In situ hybridization to whole mount embryos was performed essentially as described (23). RNA digoxigenin probes were prepared by using the Megascript Kit (Ambion, Austin, TX). Antisense and sense probes were synthesized from the cloned second exon fragment of D. sechellia.
Results
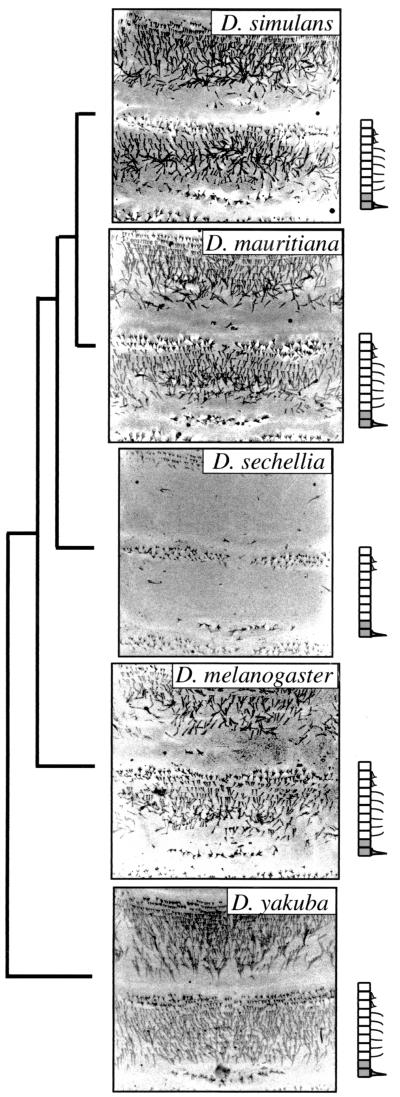
All species of the D. melanogaster species subgroup, except D. sechellia, possess a dorsal pattern of denticles and hairs similar to that described previously for D. melanogaster (24–26). The precise pattern of hairs and denticles varies between segments (25), and the following description focuses on the abdominal segments. The most anterior cells of the anterior compartment produce naked cuticle. More posteriorly, there are two to three rows of short and thick denticles and then six to eight rows of fine hairs (Fig. 1). On the lateral surface of the larvae, fine hairs are also found in the same anterior–posterior domain of each segment as the dorsal hairs (not shown). In the posterior compartment, cells of the anterior row secrete naked cuticle, and cells of the posterior row produce large thick denticles (Fig. 1).
Figure 1.
The phylogenetic distribution of dorsal hair patterns for five members of the D. melanogaster species group. A phylogeny of these species is shown (Left, modified from ref. 36). Confocal micrographs are shown for abdominal segments 1 and 2 for each species. A cartoon of the pattern of hairs is shown beside each micrograph. In the cartoons, cells in the anterior and posterior compartment of the segment are shown as white and gray rectangles, respectively. The relative positions of the three types of cuticular projections, short denticles, fine hairs, and large denticles are illustrated (see text). Anterior is up. The dorsal hair patterns for the remaining members of the group (Drosophila erecta, Drosophila orena, and Drosophila teissieri) are similar to Drosophila yakuba (not shown).
The dorsal cuticle of D. sechellia first-instar larvae differs from the above description primarily by the absence of the lawn of fine hairs both dorsally (Fig. 1) and laterally (not shown). We have noticed other minor variations in trichome patterning between species of the D. melanogaster species subgroup, but we have not characterized these in detail. Phylogenetic analysis suggests that the naked cuticle phenotype has evolved within the D. sechellia lineage (Fig. 1).
We performed a series of genetic crosses that together indicate that this phenotypic difference is caused by evolution at a single locus on the X chromosome. First, crosses between D. simulans females and D. sechellia males produced larvae with a “simulans-like” phenotype (n = 14), indicating that the D. simulans allele is completely dominant to the D. sechellia allele. No intermediate phenotypes were observed (we have not succeeded in crossing flies in the opposite direction).
Hybrid D. sechellia/D. simulans females were backcrossed to both parental species. Because recombination could occur within these hybrid females, we could estimate the number of evolved genes. The backcross to D. sechellia males produced a ratio of simulans-like to sechellia-like larvae that did not deviate significantly from a 1:1 ratio expected from the segregation of a single locus [21 simulans-like: 19 sechellia-like, χ2 = 0.10, no significant difference (n.s.)]. The backcross to D. simulans males produced a ratio of larvae that did not deviate significantly from a 1:3 ratio expected of a single sex-linked locus (52 simulans-like: 14 sechellia-like, χ2 = 0.51, n.s.).
Crosses between D. melanogaster females and D. sechellia males produced only “melanogaster-like” F1 larvae (n = 15), indicating that the D. melanogaster allele is also dominant to the D. sechellia allele [the offspring of this cross are sterile (27), preventing F2 mapping].
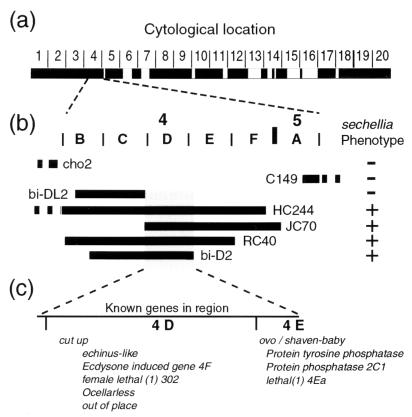
Given the dominance of the D. melanogaster allele to the D. sechellia allele, we attempted to locate the gene by performing interspecific complementation tests by using a standard set of chromosome deficiencies covering approximately 80% of the D. melanogaster X chromosome. In this test, the dominant D. melanogaster allele was hypothesized to be removed by one or several overlapping deficiencies, which would thereby reveal the recessive D. sechellia allele in a hybrid. Only Df(1)JC70 produced a proportion of “sechellia-like” larvae consistent with the deficiency having uncovered the evolved gene. Complementation tests with six additional deficiencies confirmed the initial result and refined the cytological region to 4C15-E1 (Fig. 2).
Figure 2.
Localization of the evolved gene by failure of complementation of X chromosome deficiencies. (a) The cytological regions covered by deficiencies that produced viable larvae when crossed to D. sechellia males are shown as black boxes. Approximately 75% of the chromosome was screened successfully with deficiencies. In the original screen, only Df(1)JC70 produced larvae with the D. sechellia hair pattern in the expected ratio (n = 57).[Some of the crosses yielded single larvae with a D. sechellia hair pattern. These larvae are likely the result of meiotic nondisjunction in the female parent generating nullo X eggs fertilized by X-bearing D. sechellia sperm, which in turn generated XO embryos displaying the D. sechellia hair pattern. Nondisjunction is elevated in stocks carrying balancer chromosomes and stocks with XXY females (37)]. (b) Further localization to the 4C15-E1 cytological region was performed with overlapping deficiencies. Regions deleted by deficiencies are indicated by bold horizontal lines with the name of the deficiency next to the line. The continuation of a deficiency outside this region is shown by a dashed line. Deficiencies producing larvae with a D. sechellia hair pattern in the expected frequencies when crossed to D. sechellia males are indicated with a plus sign. Deficiencies producing only larvae with a hair pattern typical of D. melanogaster are indicated by a minus sign. The distal limit of the evolved gene is defined by the right breakpoint of Df(1)bi-DL2 (4C15-D1) and the left breakpoint of Df(1) JC70 (4C15–16), and the proximal limit is defined by the right breakpoint of Df(1)bi-D2 (4D7-E1). (c) Genes known to exist within this region are listed in their approximate cytological location. The genes cut up and ovo/svb have been localized previously to the small regions, 4D1–3 and 4E1, respectively, and the three genes Protein tyrosine phosphatase 4E, Protein phosphatase 2C1, and lethal(1)4Ea have been localized previously to 4E1–2. The remaining genes shown have been localized to large regions that include 4D-E1 (see http://flybase.bio.indiana.edu for details).
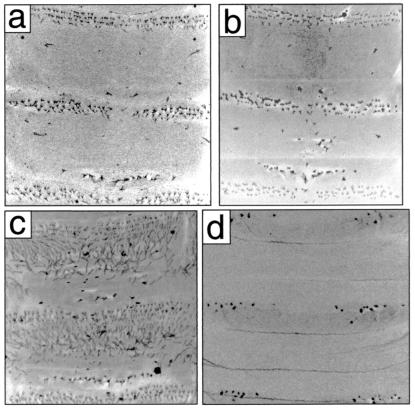
Of the candidate genes in this region (Fig. 2c), one of them, ovo/svb, had been described previously as a gene involved in denticle and hair differentiation (22, 28, 29). Mutations in this gene remove most or all of the denticles and hairs, both ventrally and dorsally, on D. melanogaster larvae (Fig. 3d).
Figure 3.
Confocal micrographs of the first and second abdominal segments of D. sechellia (a) are similar to those of a hybrid between a D. melanogaster svb1 mutant and D. sechellia (b). In contrast, a hybrid of wild-type D. melanogaster and D. sechellia (c) displays a cuticular pattern similar to D. melanogaster (see Fig. 1). The D. melanogaster svb1 mutations leads to complete loss of denticles and hairs on the dorsal surface (not shown) and loss of most denticles on the ventral surface (d). [In some svb1/D. sechellia larvae, small patches of hairs in the middle of naked cuticle were occasionally observed (b). Such hairs were never observed in hybrid backcrosses and in crosses between svb deficiencies and D. sechellia, suggesting that svb1 is not a complete loss-of-function allele.] Anterior is up.
We tested directly whether mutations of ovo/svb failed to complement the D. sechellia phenotype. The cross of svb1/FM7 and svb2/FM7 females to D. sechellia males produced embryonic cuticle patterns of three types—“melanogaster-like” (putatively, embryos carrying the D. melanogaster balancer chromosome), “sechellia-like” (putative female embryos carrying the D. sechellia X chromosome and a D. melanogaster svb mutant chromosome), and “svb-like” (putative male embryos carrying the D. melanogaster svb chromosome)—in ratios not significantly different from the expected 2:1:1 (svb1, 14:8:8, χ2 = 0.13, n.s.; svb2, 38:17:16, χ2 = 0.38, n.s.). We determined subsequently that the embryos exhibiting the svb-like phenotype were indeed males by crossing y1w1svb1 D. melanogaster heterozygous females to D. sechellia males. All “svb-like” embryonic cuticles from this cross had yellow mouthparts (n = 7), and all “sechellia-like” larvae had brown mouthparts (n = 16), confirming that the “svb-like” larvae were male and the “sechellia-like” larvae were female.
One explanation for these results is that mutations at ovo/svb generate a “sechellia-like” phenotype in any hybrid, because of hybrid incompatibilities. Crosses between D. melanogaster svb mutants and D. simulans males allow us to reject this possibility. In the cross between D. melanogaster y1w1svb1 females and D. simulans males, we observed “melanogaster-like” and “svb-like” phenotypes in the expected proportions, but never “sechellia-like” phenotypes. (One embryo was found with a trichome pattern similar to D. sechellia. This embryo also possessed extensive defects in the head, mouthparts, posterior spiracles, and Keilin's organs, as well as polarity defects in the dorsal denticles. We have not observed such pervasive defects correlated with the “sechellia-like” phenotype in other crosses, and this is probably an unrelated phenomenon.) Finally, hatched larvae from crosses between Df(1)biD2, which removes the svb region, and D. simulans males never displayed a “sechellia-like” phenotype (n = 20).
We performed a mapping experiment to determine whether the evolved gene maps near ovo-svb. Female D. simulans carrying an X chromosome marked with y1w1f2 were crossed to D. sechellia males, and the hybrid females were backcrossed to y1w1f2 D. simulans males. Two of these recessive markers are visible in the first-instar larvae; y1 produces yellow or light brown mouthparts, compared with the normal dark brown, and f2 produces thinner and more sinuous ventral denticles, similar to the phenotype of the D. melanogaster f3N allele (30), as well as stouter lateral sensory bristles. The observed number of recombinants for each class did not deviate significantly from the expectation that the evolved gene maps to the ovo/svb locus (n = 66, χ2 = 5.76, 7 degrees of freedom, n.s.).
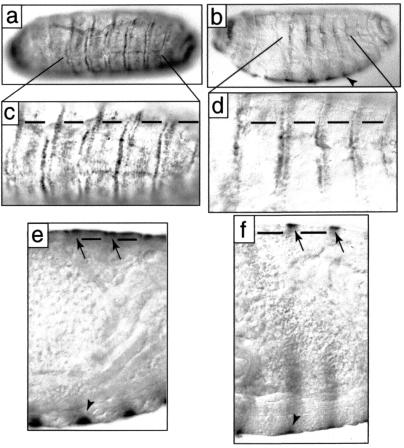
Examination of the distribution of svb transcripts indicates that cell-specific loss of svb transcription is correlated with the absence of hairs in D. sechellia (Fig. 4). The pattern of svb transcript in D. melanogaster is similar to that previously described (31). The svb transcript is detected at lower levels in the cells that differentiate the fine hairs of the dorsum, in contrast to the stronger expression detected in the rows of more robust denticles on both the dorsum and ventrum (Fig. 4 a, c, and e). In D. sechellia, the svb transcript is detected at high levels in the cells forming the ventral denticle rows and in other trichome-forming cells, including the dorsal denticle rows (Fig. 4 b, d, and f). The svb transcript was not detected in the dorsal cells that differentiate naked cuticle (Fig. 4 b, d, and f).
Figure 4.
The pattern of svb expression in D. melanogaster (a, c, and e) and D. sechellia (b, d, and f) stage 17 embryos. Dorsal views are shown in a, c, and d, dorso–lateral in b, and lateral optical cross sections in e and f. The svb transcript is detected in both species in rows corresponding to the dorsal stout denticles (a–d and arrows in e and f). svb transcript is detected at lower levels in the rows giving rise to fine hairs in D. melanogaster (horizontal bars in c and e) and is not detected in the corresponding positions in D. sechellia (horizontal bars in d and f). The cells differentiating the ventral denticle belts express high levels of svb (arrowheads in b, e, and f). Anterior is left in all images, and dorsal is up in b, e, and f.
Discussion
We have shown that a dramatic difference in larval morphology between sibling species is caused by evolution at a single locus. Complementation tests and mapping experiments indicate that evolution at the ovo/svb locus is responsible for the difference in dorsal hair patterning between D. sechellia and other species of the D. melanogaster species subgroup. This change may be caused by evolution of cis-regulatory transcriptional control of ovo/svb.
The regulatory nature of the change is supported by several observations. First, independent regulatory elements drive the ovo and svb functions (22, 31, 32). The ovo function is required for development of the female germline, and ovo mutants can be rescued by genomic DNA spanning the known exons of ovo. The svb function is required for the differentiation of denticles and hairs in the larvae. Some svb mutants, including breakpoints 5′ of the known exons of ovo/svb, are not rescued by the ovo transgene, suggesting that regulatory elements 5′ of the known exons are required for svb function (22). Second, the pattern of naked cuticle and denticles in D. sechellia is not identical to the svb loss-of-function phenotype (28) and instead appears to represent the loss of a subset of the hairs lost in svb mutants (Fig. 3). This suggests that the transition to the D. sechellia phenotype was caused by cis-regulatory evolution of part of svb function. Our analysis of the distribution of ovo/svb transcripts in embryos of D. melanogaster and D. sechellia (Fig. 4) suggests that this change occurred at the level of transcriptional control of ovo/svb.
Modular enhancer architecture (33) allows mutations to alter only one part of a gene's function, without generating multiple deleterious pleiotropic effects. In the case we have documented, evolution of ovo/svb resulted in an alteration in the function of only part of ovo/svb function, apparently without pleiotropic effects on other aspects of trichome and hair patterning. Because the ovo function is also required for female germline differentiation, variation at this locus might be expected to affect also ovarian development. In fact, females of D. sechellia have fewer ovarioles than females of its sibling species. However, none of the genes generating this difference map near ovo/svb (34). Therefore, evolution of the cis-regulatory regions of patterning genes can result in relatively dramatic evolutionary transitions without potentially deleterious pleiotropic consequences, even when such genes play pleiotropic roles in development.
Models of adaptation suggest that single mutational events causing dramatic phenotypic alterations are more likely to be fixed by strong selection and at the beginning of a bout of adaptation (6, 35). This contrasts with the traditional Darwinian view that large differences arise from the accumulation of many small changes. Although our experiments indicate that a major difference in larval hair patterning is caused by evolution of a single gene, this large change may have resulted from the accumulation of multiple mutations of smaller effect within the cis-regulatory region of ovo/svb. In either case, the results are surprising, but for different reasons. If this morphological transition is caused by a single mutational change, then mutations of relatively large effect must be recognized as contributors to species differences. If, however, the transition is caused by multiple mutations at ovo/svb, then we must explain why all of the mutations occurred at a single locus and were not distributed among multiple loci. The latter result would support the idea that a limited number of genes may be available to generate evolution of at least some morphological features. Resolution of this problem requires experiments to identify the individual mutations at ovo/svb that have generated these differences. Given the complexity of the ovo/svb locus and particularly the currently unknown structure of the svb regulatory regions, these experiments will not be trivial, but they are tractable in Drosophila, and they are currently under way.
This study demonstrates further the power of using species closely related to the “model” species, D. melanogaster, for studying problems in evolution and development. Only the powerful tools and vast knowledge of D. melanogaster allowed us to move quickly from observation of morphological differences to study of a single gene. The D. melanogaster species group displays a diversity of morphology, behavior, physiology, and life history, and all of these traits are amenable to comparatively thorough genetic analysis because of the phylogenetic proximity of D. melanogaster. Further studies of the D. melanogaster species group will generate central insights into the process, as well as the pattern, of developmental evolution.
Acknowledgments
For fly stocks, we thank John Roote at the Ashburner lab (Cambridge, U.K.), the Nüsslein–Volhard lab, the Bloomington Stock Center, and the Species Stock Center. We thank A. Orr for suggesting a useful control experiment and Christen Mirth, the editor, and anonymous referees for useful comments on an earlier draft. This work was funded by a Programa Gulbenkian de Doutoramento em Biologia e Medicina and Praxis XXI Fellowship (Portugal) to E.S. and a Biotechnology and Biological Sciences Research Council David Phillips Research Fellowship to D.L.S.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: n.s., no significant difference; ovo/svb, ovo/shaven-baby.
References
- 1.Averof M, Akam M. Nature (London) 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]
- 2.Carroll S B, Weatherbee S D, Langeland J A. Nature (London) 1995;375:58–61. doi: 10.1038/375058a0. [DOI] [PubMed] [Google Scholar]
- 3.Panganiban G, Sebring A, Nagy L, Carroll S. Science. 1995;270:1363–1366. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- 4.Gellon G, McGinnis W. BioEssays. 1998;20:116–125. doi: 10.1002/(SICI)1521-1878(199802)20:2<116::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 5.Fisher R A. The Genetical Theory of Natural Selection. NY: Dover; 1930. pp. 1–188. [Google Scholar]
- 6.Orr H A. Evolution (Lawrence, KS) 1998;52:935–949. doi: 10.1111/j.1558-5646.1998.tb01823.x. [DOI] [PubMed] [Google Scholar]
- 7.True J R, Liu J, Stam L F, Zeng Z-B, Laurie C C. Evolution (Lawrence, KS) 1997;51:816–832. doi: 10.1111/j.1558-5646.1997.tb03664.x. [DOI] [PubMed] [Google Scholar]
- 8.Zeng Z, Liu J, Stam L F, Kao C, Mercer J M, Laurie C C. Genetics. 2000;154:299–310. doi: 10.1093/genetics/154.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pugh A R, Ritchie M G. Heredity. 1996;77:378–382. doi: 10.1038/hdy.1996.156. [DOI] [PubMed] [Google Scholar]
- 10.Palopoli M F, Wu C-I. Genetics. 1994;138:329–341. doi: 10.1093/genetics/138.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyne J A. Genet Res. 1996;68:211–220. doi: 10.1017/s0016672300034182. [DOI] [PubMed] [Google Scholar]
- 12.Jones C D. Genetics. 1998;149:1899–1908. doi: 10.1093/genetics/149.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carson H L, Val F C, Templeton A R. Proc Natl Acad Sc USA. 1994;91:6315–6318. doi: 10.1073/pnas.91.14.6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osborne K A, Robichon A, Burgess E, Butland S, Shaw R A, Coulthard A, Pereira H S, Greenspan R J, Sokolowski M B. Science. 1997;277:834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- 15.ffrench-Constant R H, Rocheleau T A, Steichen J C, Chalmers A E. Nature (London) 1993;363:449–451. doi: 10.1038/363449a0. [DOI] [PubMed] [Google Scholar]
- 16.Doebley J, Stec A, Hubbard L. Nature (London) 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 17.Dickinson W J, Tang Y, Schuske K, Akam M. Evolution (Lawrence, KS) 1993;47:1396–1406. doi: 10.1111/j.1558-5646.1993.tb02162.x. [DOI] [PubMed] [Google Scholar]
- 18.Stern D L, Sucena E. In: Drosophila: A Laboratory Manual. Ashburner M, Hawley S, Sullivan B, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 2000. pp. 601–615. [Google Scholar]
- 19.Coyne J A, Charlesworth B. Genetics. 1997;145:1015–1030. doi: 10.1093/genetics/145.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R'kha S, Capy P, David J R. Proc Natl Acad Sci USA. 1991;88:1835–1839. doi: 10.1073/pnas.88.5.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Mével-Ninio M, Terracol R, Kafatos F C. EMBO J. 1991;10:2259–2266. doi: 10.1002/j.1460-2075.1991.tb07762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 24.Martinez Arias A. In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 517–608. [Google Scholar]
- 25.Kuhn D T, Sawyer M, Ventimiglia J, Sprey Th E. Drosophila Information Service. 1992;71:218–222. [Google Scholar]
- 26.Bokor P, DiNardo S. Development (Cambridge, UK) 1996;122:1083–1092. doi: 10.1242/dev.122.4.1083. [DOI] [PubMed] [Google Scholar]
- 27.Lemeunier F, Ashburner M. Proc R Soc London Ser B. 1976;193:275–294. doi: 10.1098/rspb.1976.0046. [DOI] [PubMed] [Google Scholar]
- 28.Wieschaus E, Nusslein-Volhard C, Jurgens G. Roux's Arch Dev Biol. 1984;193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- 29.Payre F, Vincent A, Carreno S. Nature (London) 1999;400:271–275. doi: 10.1038/22330. [DOI] [PubMed] [Google Scholar]
- 30.Dickinson W J, Thatcher J W. Cell Motil Cytoskeleton. 1997;38:9–21. doi: 10.1002/(SICI)1097-0169(1997)38:1<9::AID-CM2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Mével-Ninio M, Terracol R, Salles C, Vincent A, Payre F. Mech Dev. 1995;49:83–95. doi: 10.1016/0925-4773(94)00305-7. [DOI] [PubMed] [Google Scholar]
- 32.Mével-Ninio M, Mariol M-C, Gans M. EMBO J. 1989;8:1549–1558. doi: 10.1002/j.1460-2075.1989.tb03539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnone M I, Davidson E H. Development (Cambridge, UK) 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- 34.Coyne J A, Rux J, David J R. Genet Res. 1991;57:113–122. doi: 10.1017/s0016672300029177. [DOI] [PubMed] [Google Scholar]
- 35.Lande R. Heredity. 1983;50:47–65. [Google Scholar]
- 36.Harr B, Weiss S, David J R, Brem G, Schlötterer C. Curr Biol. 1998;8:1183–1186. doi: 10.1016/s0960-9822(07)00490-3. [DOI] [PubMed] [Google Scholar]
- 37.Ashburner M. Drosophila: A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]