Abstract
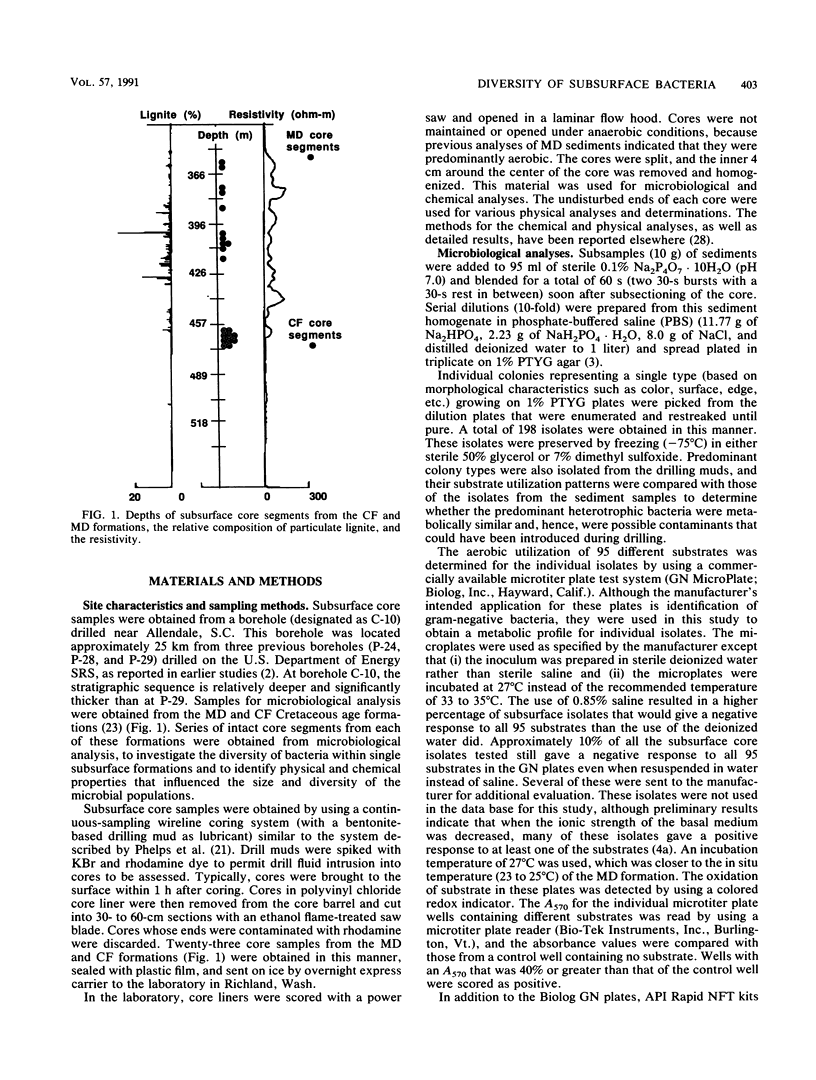
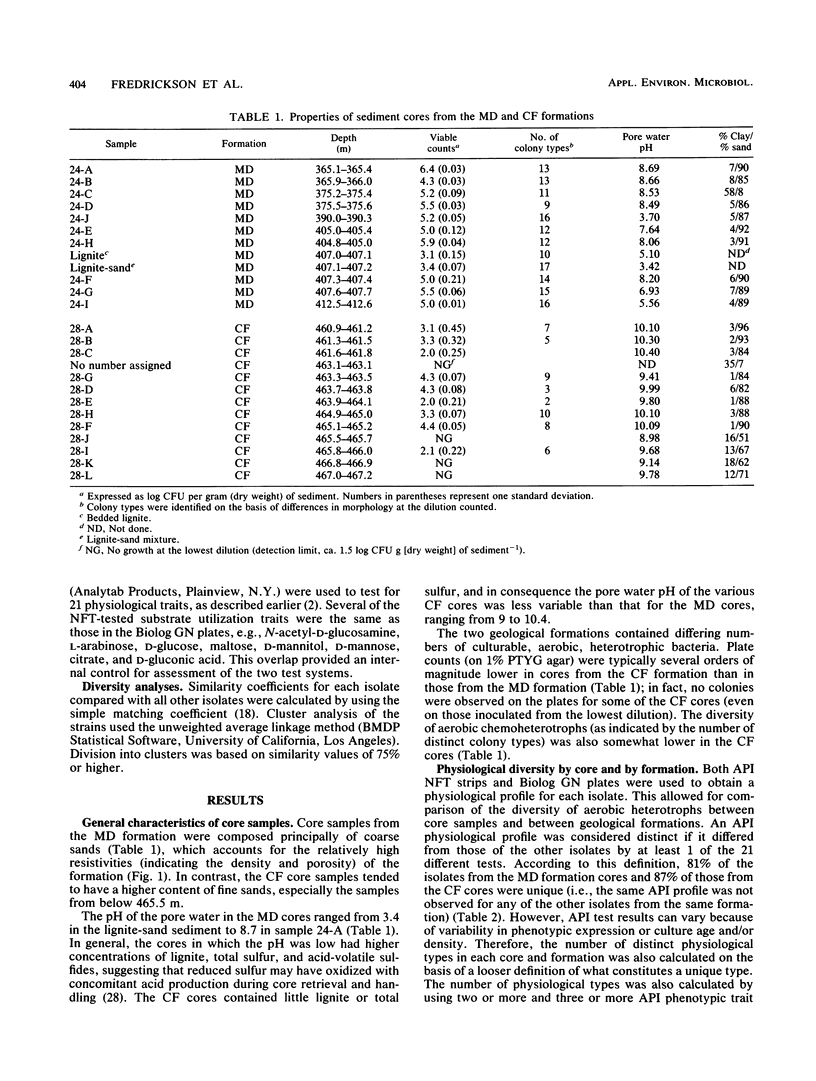
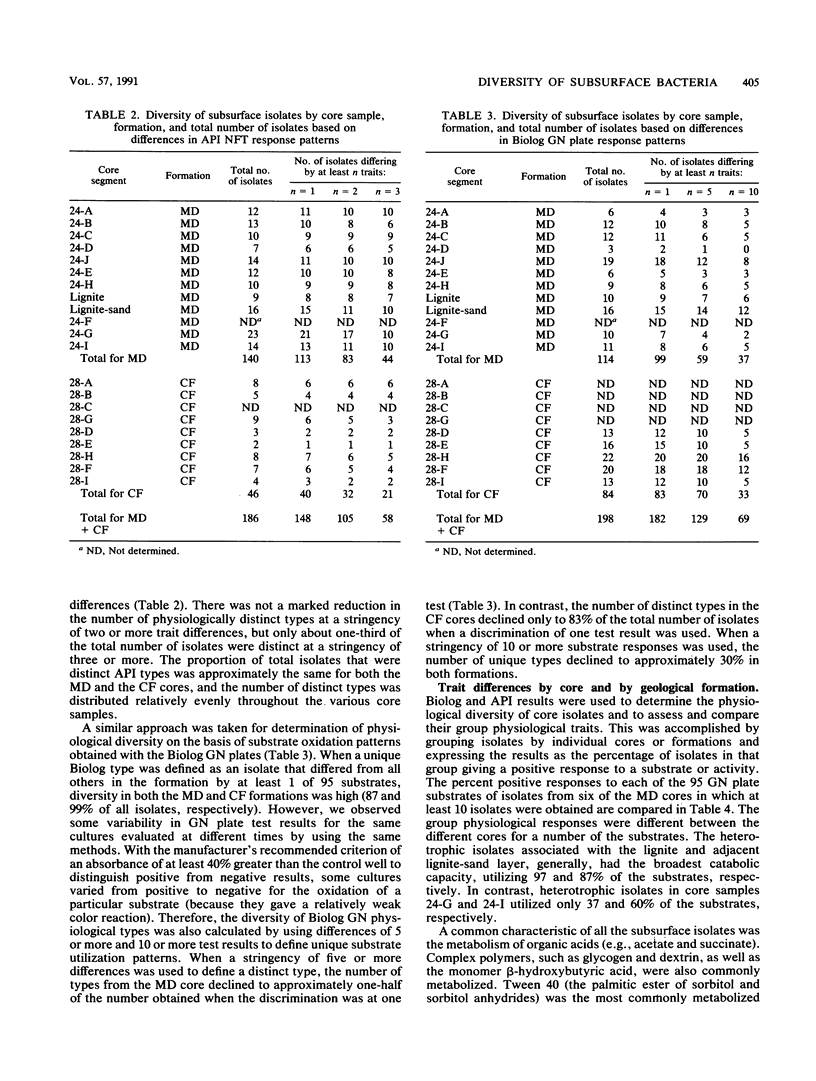
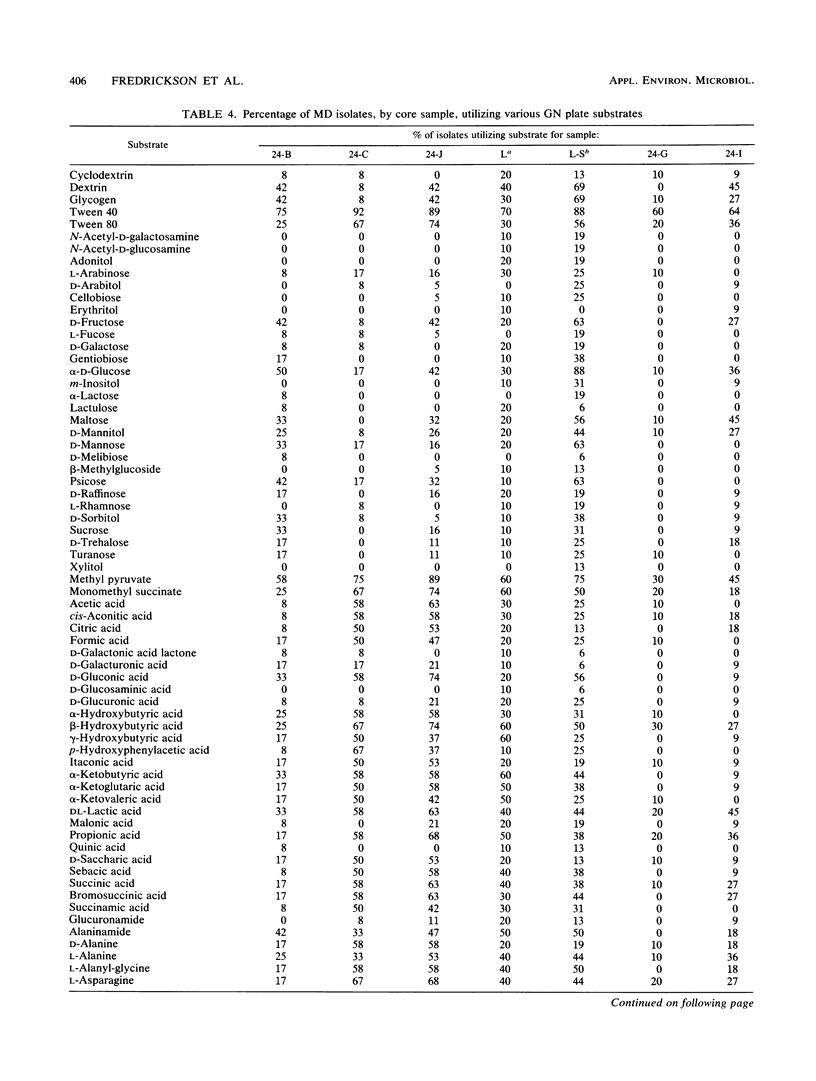
A series of 23 intact core segments was obtained from two distinct deep subsurface geological formations, the Middendorf and the Cape Fear formations, underlying the southeastern coastal plain of South Carolina. The Middendorf formation in this region consists of permeable, saturated, sandy sediments; the Cape Fear formation consists mainly of less permeable sediments. The core segments were separated by vertical distances ranging from several centimeters to 48 m. Aerobic chemoheterotrophic bacteria were enumerated on a dilute medium, and populations ranged from 3.1 to 6.4 log CFU g of sediment-1 in the Middendorf cores and from below detection to 4.3 log CFU g-1 in the Cape Fear cores. A total of 198 morphologically distinct colony types were isolated, purified, and subjected to 108 different physiological measurements. The isolates from the two formations were distinct (i.e., they produced substantially different response patterns to the various physiological measurements), as were those in different core samples from the same formation. Cluster analysis revealed 21 different biotypes based on similarities of 75% or higher in response patterns to 21 physiological assays. One biotype contained 57 (29%) of the subsurface isolates, 10 biotypes contained 5 or more isolates, and the remainder had 4 or fewer. The organic compounds that were most commonly metabolized by the subsurface bacteria included Tween 40 (85%) and β-hydroxybutyric acid (60%). Organic acids, in general, were also commonly metabolized by the subsurface bacteria. Isolates from the Cape Fear core segments were capable of metabolizing a higher percentage of the substrates than were bacteria isolated from the Middendorf formation. Although the heterogeneous distributions of bacteria in deep subsurface sediments may make it difficult to use aquifer microcosms to predict in situ biotransformation rates, the diversity of the physiological properties of these organisms offers promise for in situ remediation of contaminants.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balkwill D. L., Fredrickson J. K., Thomas J. M. Vertical and horizontal variations in the physiological diversity of the aerobic chemoheterotrophic bacterial microflora in deep southeast coastal plain subsurface sediments. Appl Environ Microbiol. 1989 May;55(5):1058–1065. doi: 10.1128/aem.55.5.1058-1065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill D. L., Ghiorse W. C. Characterization of subsurface bacteria associated with two shallow aquifers in oklahoma. Appl Environ Microbiol. 1985 Sep;50(3):580–588. doi: 10.1128/aem.50.3.580-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman F. J., Denovan B. A., Hicks R. J., Fredrickson J. K. Isolation and characterization of quinoline-degrading bacteria from subsurface sediments. Appl Environ Microbiol. 1989 Apr;55(4):1029–1032. doi: 10.1128/aem.55.4.1029-1032.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapelle F. H., Lovley D. R. Rates of microbial metabolism in deep coastal plain aquifers. Appl Environ Microbiol. 1990 Jun;56(6):1865–1874. doi: 10.1128/aem.56.6.1865-1874.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiorse W. C., Wilson J. T. Microbial ecology of the terrestrial subsurface. Adv Appl Microbiol. 1988;33:107–172. doi: 10.1016/s0065-2164(08)70206-5. [DOI] [PubMed] [Google Scholar]
- Torsvik V., Salte K., Sørheim R., Goksøyr J. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl Environ Microbiol. 1990 Mar;56(3):776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]



