Abstract
In crystals of the key respiratory and photosynthetic electron transfer protein called ubihydroquinone:cytochrome (cyt) c oxidoreductase or cyt bc1, the extrinsic [2Fe2S] cluster domain of its Fe-S subunit assumes several conformations, suggesting that it may move during catalysis. Herein, using Rhodobacter capsulatus mutants that have modifications in the hinge region of this subunit, we were able to reveal this motion kinetically. Thus, the bc1 complex (and possibly the homologous b6f complex in chloroplasts) employs the [2Fe2S] cluster domain as a device to shuttle electrons from ubihydroquinone to cyt c1 (or cyt f). We demonstrate that this domain movement is essential for cyt bc1 function, because a mutant enzyme with a nonmoving Fe-S subunit has no catalytic activity, and one with a slower movement has lower activity. This motion is apparently designed with a natural frequency slow enough to assure productive Qo site charge separation but fast enough not to be rate limiting. These findings add the unprecedented function of intracomplex electron shuttling to large-scale domain motions in proteins and may well provide a target for cyt bc1 antibiotics.
Keywords: Rhodobacter capsulatus, photosynthetic and respiratory electron transfer, mitochondrial complex III, protein domain motion, Rieske Fe-S subunit
When different crystal structures reveal dramatically different protein conformations, large amplitude domain movements are often inferred. However, in only a few cases such as myosin (1), flagellar motor (2), and ATP synthase (3, 4) have such movements been visualized. The cytochrome (cyt) bc1 (or its cyt b6f counterpart in chloroplasts) is a key component of respiratory and photosynthetic electron transfer chains (5, 6). Recent crystal structures of the mitochondrial cyt bc1 have revealed that the extrinsic [2Fe2S] cluster domain of the Fe-S subunit occupies various locations within this enzyme complex (7–10). It has been observed in either a position proximal to the ubihydroquinone (QH2) oxidation catalytic site (Qo position) from which it takes electrons or a position close to cyt c1 subunit (c1 position) to which it donates electrons (Fig. 1). Because of the large distances observed between the electron-donating and electron-accepting cofactors of the cyt bc1 in the different structures, no one of these locations can support sufficiently rapid electron tunneling (11) to meet the observed turnover rates (12, 13) and the specific substrate–product interactions (14) that occur at the QH2 oxidation site. Thus, an unprecedented intracomplex electron shuttle motion to transfer electrons during catalysis has been suggested (8). However, neither the presumably essential movement nor the electron transfer associated with it has been visualized before this work.
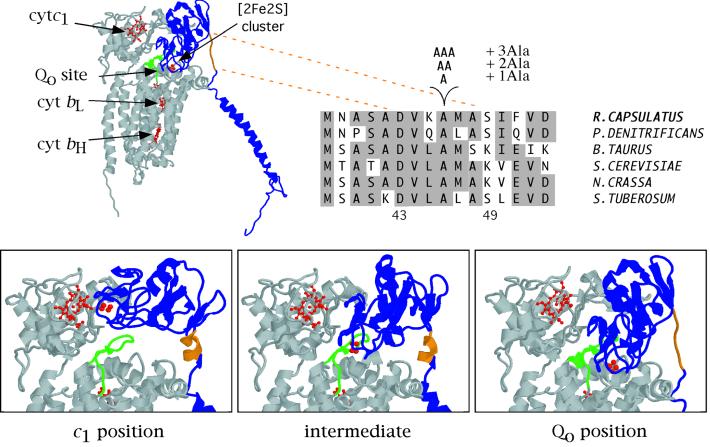
Figure 1.
Different positions of the Fe-S subunit extrinsic domain and location of the alanine residue insertions on the linker region. The structure of the three catalytic subunits of the bovine heart cyt bc1 as well as a close view of the bovine Fe-S subunit in two different crystal forms (ref. 9; c1 and intermediate positions) and that of the chicken heart in the presence of stigmatellin (ref. 8; Qo position) are shown. The Fe-S subunit is in blue with its flexible linker region encompassing the amino acid residues 67–73 (corresponding to 43–49 in Rhodobacter capsulatus) in orange. Cyt b and cyt c1 subunits are in gray, and the cofactors hemes bL, bH, and c1 and the [2Fe2S] cluster are in red, with part (residues 255–270 in bovine numbering) of the ef loop of cyt b in green. The Fe-S subunit sequences from a few selected species (Paracoccus denitrificans, Bos taurus, Saccharomyces cerevisiae, Neurospora crassa, and Solanum tuberosum) aligned with the sequence of R. capsulatus between the positions 38 and 53 and the position of the alanine residues insertion site are also shown, with identical residues highlighted in gray.
In light-activated energy transduction systems, such as the one provided by the photosynthetic bacterium R. capsulatus, a short flash of light (<10-μs duration) can activate the photochemical reaction center, thereby inducing oxidation of two equivalents of cyt c (c1 and a mixture of c2/cy) that are presented to each cyt bc1 in about 50 μs (12, 13). The cyt bc1 is then primed to complete its catalytic cycle (Fig. 2 Left A and B). Extensive electron paramagnetic resonance (EPR) spectroscopy data establish that the equilibrium position of the reduced [2Fe2S] cluster domain is located at the Qo position (14). Both the amount and rate of flash-oxidized cyt c reduction depend on the arrival of electrons via the initially reduced [2Fe2S] cluster and, later, from the millisecond oxidation of QH2 at the Qo site (12, 13). Fig. 2 Left C–H depicts how the electrons might get from the Qo site to cyt c1 if brought about by the movement of the [2Fe2S] cluster domain.
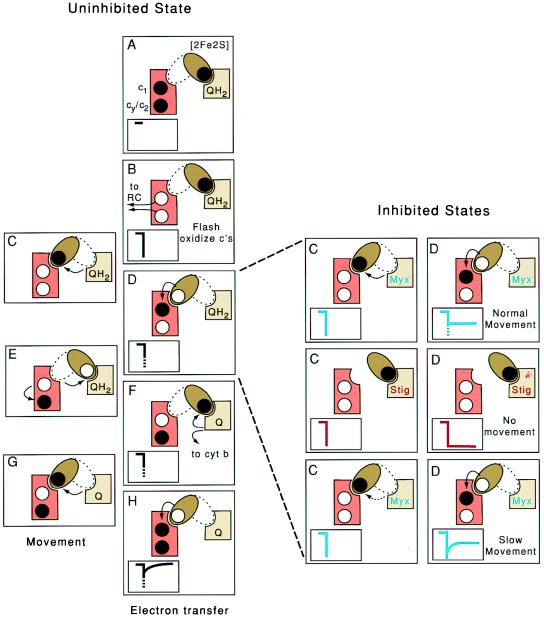
Figure 2.
Proposed intracomplex electron shuttle mechanism in cyt bc1. (Left) Schematic representations of the different steps of electron transfer (A, B, D, F, and H) and movement (C, E, and G) in the uninhibited cyt bc1. The [2Fe2S] cluster domain, the cyt c complement (c1 + cy/c2) and the Qo site are shown as a brown ellipsoid, orange rectangle, and tan square, respectively. Black dots represent electrons located on the [2Fe2S] cluster and the cyt c heme complement before flash oxidation (A). Arrows refer to electron transfers between cofactors and movement of the [2Fe2S] cluster domain between its Qo and c1 positions. (Insets) Kinetics of cyt c oxidation (downward deflection) and cyt c reduction (upward deflection). The dotted parts of the traces in D, F, and H indicate the predicted unresolved fast phase. On flash activation of the reaction center and oxidation of the cyt c complement (B), the [2Fe2S] cluster domain is proposed to move from its Qo to c1 position (C), transfer an electron to cyt c1 of the cyt c complement (D), and return to the Qo position (E). Then, QH2 is oxidized to ubiquinone (Q); one electron is transferred to the [2Fe2S] cluster; and the other is transferred to the low potential cyt b chain (F). The [2Fe2S] cluster domain moves back to the c1 position (G) where it transfers another electron (H). (Right) Representation of the steps C (movement) and D (electron transfer) shown in Left in inhibited states. In the presence of the Qo site inhibitor myxothiazol (Top), normal movement of the [2Fe2S] cluster domain will yield partial reduction of the cyt c complement with an unresolved fast phase (blue traces). In the presence of stigmatellin or in a mutant locking the [2Fe2S] cluster domain in Qo position (Middle), the cyt c complement should remain completely oxidized (red traces), whereas in a mutant slowing the movement (Bottom), this fast phase should be time-resolved (blue traces).
Fig. 2 Right also shows the effects of two powerful natural antibiotics, myxothiazol and stigmatellin, that are commonly used in the laboratory to inhibit the Qo site reaction in quite different ways. When myxothiazol is present, no QH2 oxidation occurs, simply because myxothiazol displaces QH2. In this case, cyt c reduction kinetics suggest that the initially reduced [2Fe2S] cluster can still move from the Qo site to the cyt c1 position and contribute to cyt c reduction, which is due only partially to their similar redox potentials. However, this electron transfer from the [2Fe2S] cluster to cyt c1 heme, which is now independent of Qo site catalysis, has never been observed (Fig. 2 Right Top C and D). Thus, if the [2Fe2S] domain moves as proposed, it must occur on a time scale so as to be hidden within the 50-μs envelope required to activate the entire system. When stigmatellin is added, the [2Fe2S] cluster domain is trapped in the Qo position, as evidenced by EPR (15), crystallographic (8), and reconstitution (16) data. Under these conditions, no electron transfer takes place from the [2Fe2S] cluster to cyt c1 heme, leaving the cyt c complement fully oxidized (Fig. 2 Right Middle C and D). A critical test of the model would be to devise ways to impede the movement sufficient enough to bring it out of the 50-μs envelope but not to create a situation as encountered with stigmatellin, where it is stopped altogether. Only then will the reduction of the flash-oxidized cyt c visibly depend on the rate of this movement and be strictly controlled by it in the presence of myxothiazol, as shown in Fig. 2 Right Bottom C and D.
Comparison of various crystallographic data (7–10) indicates that movement of the [2Fe2S] cluster domain would require conformational changes of the linker region encompassing amino acid residues 67–73 in the bovine sequence (corresponding to 43–49 in R. capsulatus numbering) and connecting its fixed hydrophobic anchor and extrinsic carboxyl-terminal portions (Fig. 1). We considered that this putative hinge region might therefore be vulnerable to mutations that could obstruct this mobility. Herein, using R. capsulatus mutants with alanine residue insertions, we were able to reveal this motion kinetically and demonstrate that it is essential for the function of the cyt bc1.
Materials and Methods
Bacterial Strains and Growth Conditions.
Escherichia coli or R. capsulatus strains were grown as described in ref. 17 in Luria–Bertani broth or mineral-peptone-yeast-extract enriched medium, respectively, and in the presence of appropriate antibiotics. Respiratory or photosynthetic growth of R. capsulatus strains was at 35°C in the dark under semiaerobic conditions or in anaerobiosis under continuous light, respectively. MT-RBC1 is a bc1− strain in which the chromosomal copy of the petABC operon has been deleted and replaced by a gene cartridge conferring resistance to spectinomycin (17). The strain pMTS1/MT-RBC1 corresponds to MT-RBC1 complemented in trans with the plasmid pMTS1, which provides resistance to kanamycin and contains a wild-type copy of petABC.
Molecular Genetic Techniques.
Engineering of mutations located in the linker region of the Fe-S subunit was facilitated by the creation of a unique MluI restriction site in pMTS1 via a silent mutation at its codon Ala-40, yielding plasmid pMTS1-MluI (E.D., M.V.-V., and F.D., unpublished work). The alanine insertion mutations were created by PCR with an upstream common primer (5′-GTCTTGGGCTCGTAA-3′) and the following downstream specific primers: +1Ala, 5′-ATGAACGCGTCGGCCGACGTCAAGGCGGCGATGGC- ATCGATCTTCG-3′; +2Ala, 5′-ATGAACGCGTCGGCCGACGTCAAGGCGGCGGCGATGGCATCGATCTTCG-3′; +3Ala, 5′-ATGAACGCGTCGGCCGACGTCAAGGCGGCGGCGGCGATGGCATCGATCTTCG-3′. The MluI–ApaLI fragment containing the mutation thus generated was then exchanged with its wild-type counterpart in pMTS1-MluI, and the newly constructed plasmids were introduced into MT-RBC1 via triparental crosses (17). In all cases, the presence of the desired mutation and absence of any additional mutation on the insert thus exchanged was confirmed by DNA sequencing.
Biochemical and Biophysical Techniques.
Chromatophore membrane preparation, protein determination, and 2,3-dimethoxy-5-methyl-6-decyl-1,4-benzohydroquinone:cyt c reductase assays were performed as described (17). SDS/PAGE was performed by using an acrylamide concentration of 15% (wt/vol), and gels were stained with Coomassie blue. Immunoblot analyses were performed as described (17), with monoclonal or polyclonal antibodies specific for R. capsulatus cyt bc1, which was purified as described (18). Proteolysis experiments with thermolysin were done at room temperature in 50 mM Tris⋅HCl (pH 8.0) containing 100 mM NaCl, 0.01% dodecyl maltoside, and 20 μM stigmatellin, when specified (M.V.-V., E.D., C. R. Moomaw, C. A. Slaughter, and F.D., unpublished work). Aliquots were analyzed by immunoblotting with polyclonal antibodies against the Fe-S protein of R. capsulatus.
Light-induced, single-turnover, time-resolved kinetics were performed as described (19) by using chromatophore membranes and a single wavelength spectrophotometer (Biomedical Instrumentation Group, University of Pennsylvania) in the presence of 2.5 μM valinomycin, N-ethyl-dibenzopyrazine ethyl sulfate, N-methyl-dibenzopyrazine methyl sulfate, 2,3,5,6-tetramethyl-1,4-phenylenediamine, and 2-hydroxy-1,4-naphthoquinone. Transient cyt c reduction kinetics initiated by a short saturating flash (8 μs) from a xenon lamp were followed at 550–540 nm, and cyt b reduction was followed in the presence of antimycin at 560–570 nm. The concentrations of antimycin, myxothiazol, and stigmatellin used were 5, 5, and 1 μM, respectively, and the ambient potential was poised at 100, 200, or 400 mV as indicated.
Oxidative titrations of the Fe-S subunit [2Fe2S] cluster in chromatophore membranes were conducted potentiometrically according to Dutton (20) in the presence of 100 μM tetrachlorohydroquinone, 2,3,5,6-tetramethyl-1,4-phenylenediamine, 1,2-naphthoquinone-4-sulfonate, 1,2-naphthoquinone, N-ethyl-dibenzopyrazine ethyl sulfate, and N-methyl-dibenzopyrazine methyl sulfate, and in the presence of 100 μM stigmatellin or 300 μM myxothiazol when indicated. EPR spectroscopy of these samples was performed as described (21) by using a Bruker (Billerica, MA) ESP-300E, equipped with an Oxford Instruments (Oxon, England) ESR-9 helium cryostat, under the following conditions: sample temperature, 20 K; microwave power, 2 mW; modulation amplitude, 20.243 G; modulation frequency, 100 kHz; microwave frequency, 9.45 GHz.
Chemicals.
All chemicals were as described (22).
Results and Discussion
The Alanine Insertion Mutants and Their Initial Characterization.
In an attempt to interfere with the function of the Fe-S subunit hinge region (corresponding to residues 43–49 in R. capsulatus), mutants with insertions of one (+1Ala), two (+2Ala), or three (+3Ala) alanine residues were engineered between position 46 and 47 (corresponding to residues 70 and 71 in bovine numbering; Fig. 1). These mutants properly assembled their cyt bc1, as shown by immunoblot analyses of the subunits and spectroscopic quantification of their cofactors (b- and c-type hemes, [2Fe2S] cluster). The [2Fe2S] cluster–Qo site interactions of the mutants were normal, and their [2Fe2S] cluster domains were located in the Qo position when reduced, as indicated by the position and lines shapes of their EPR gx signals (14) in the absence and presence of stigmatellin (data not shown). However, the +2Ala and +3Ala mutants were unable to grow, and the +1Ala mutant grew poorly under photosynthetic growth conditions. This finding indicated a cyt bc1 defect, because in phototrophic bacteria like R. capsulatus, this enzyme is required for cyclic electron transport. In agreement with these results, the cyt bc1 of the +1Ala mutant was poorly functional, and the cyt bc1 of the +2Ala or +3Ala mutants was nonfunctional as revealed by their single turnover activities (Table 1). Surprisingly, the steady-state activity of the +1Ala mutant was abnormally high in detergent-dispersed membranes.
Table 1.
Characteristics of R. capsulatus mutants
| Strains | Ps phenotype* | Steady-state activity, %† | Electron transfer QH2 → cytc, %‡ | Em7 [2Fe-2S], mV§ |
|---|---|---|---|---|
| Wild-type | Ps+ | 100 | 100 | 310 |
| +1Ala | Psslow | 120 | 35 | 370 |
| +2Ala | Ps− | 5 | 2 | 410 |
| +3Ala | Ps− | 2 | 2 | nd |
| A46T¶ | Ps+ | 70 | 65 | 386 |
| Y147A∥ | Ps− | 13 | 6 | 310 |
Ps+ and Ps− indicate photosynthetic competence and incompetence, respectively.
† Steady-state bc1 complex activity was determined by measuring the 2,3-dimethoxy-5-methyl-6-decyl-1,4-benzohydroquinone:cyt c reductase activity (17) and is expressed as a percentage of the wild-type activity, which was, in this particular instance, 3.4 μmol cyt c reduced min−1⋅mg of membrane protein−1.
‡ QH2 to cyt c electron transfer rates were determined by recording cyt c rereduction kinetics at 550–540 nm and fitting them to a single exponential equation (22). The rates are expressed as a percentage of the wild-type rate, which was 300 s−1, and reflect single turnover bc1 complex activity. Note that under these conditions, the electron transfer activities from QH2 → cyt b are not significantly different than those from QH2 → cyt c shown here.
§ The Em7 values were obtained after fitting the amplitude of the EPR gy signal during potentiometric titration of the [2Fe-2S] cluster as described in Materials and Methods. nd, not determined.
¶ A 46 T mutation is located in the Fe-S subunit, and the data are taken from ref. 21.
∥Y147A mutation is located in cyt b subunit, and the data are taken from ref. 23.
The +2Ala Mutant Contains a Nonmoving Fe-S Subunit.
Further insights toward the effects of the alanine insertion mutations were gained by analyzing their cyt c reduction kinetics in the presence of the Qo site inhibitors stigmatellin and myxothiazol. In the case of the +2Ala (Fig. 3B) and +3Ala (data not shown) mutants, cyt c reduction kinetics without inhibitor or with myxothiazol or stigmatellin were almost identical to those observed with a stigmatellin-inhibited native enzyme (Fig. 3A). Thus, this absence of electron transfer from the [2Fe2S] cluster to cyt c1 heme, especially in the presence of myxothiazol, under which conditions this event is independent of Qo site catalysis, suggested that in these mutants the [2Fe2S] cluster domain did not move during the 100-ms time scale of the measurements. It is noteworthy that these mutants do not resemble the Qo site-inactive mutants such as Y147A (Table 1) (23). The latter mutants behaved even in the absence of any inhibitor as if they had already been inhibited by myxothiazol, but they could still transfer electrons from the [2Fe2S] cluster to cyt c1 heme as indicated by their cyt c reduction kinetics in the presence of stigmatellin (Fig. 3D).
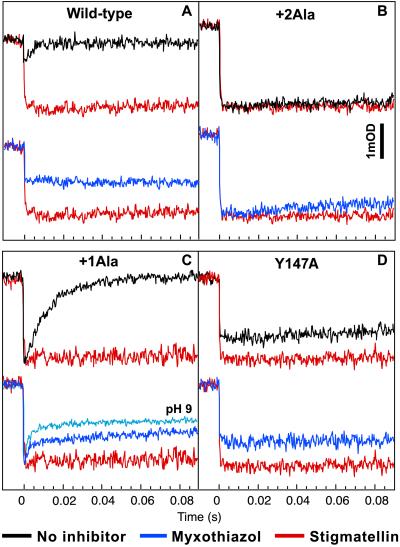
Figure 3.
Flash-oxidized cyt c reduction kinetics in wild type, +2Ala, and +1Ala mutants and Qo site-defective mutant Y147A. The traces obtained with no inhibitor (black), myxothiazol (blue), or stigmatellin (red) with wild type (A), +2Ala mutant (B), +1Ala mutant (C), and Y147A (D) are shown. The ambient redox potential was poised at 100 mV at pH 7.0 such that the Q pool contained substrate QH2 or at 200 mV for Y147A. Transient kinetics of the flash-induced oxidation and reduction of the cyt c complement were followed at 550–540 nm. The concentrations of myxothiazol and stigmatellin were 5 and 1 μM, respectively, for full inhibition. In C, the traces obtained with myxothiazol at pH 9.0 (light blue), under which conditions a lower [2Fe2S] cluster redox midpoint potential contributes, as expected, to a more prominent reduction of cyt c, are also shown.
Additional support for the suggestion that the addition of two alanine residues to the hinge region of the Fe-S subunit has locked its extrinsic domain in the Qo position included the presence of a higher amount of Q trapped in the Qo site of the purified +2Ala mutant cyt bc1 (EPR spectra not shown) and a higher Em of the [2Fe2S] cluster (ΔEm = +100 mV; Table 1). These indicate a stronger interaction of the [2Fe2S] cluster domain with the Qo site, of the kind observed in the presence of stigmatellin. Furthermore, the +2Ala mutant Fe-S subunit showed resistance to thermolysin-mediated, conformation-sensitive proteolysis (ref. 24 and M.V.-V., E.D., C. R. Moomaw, C. A. Slaughter, and F.D., unpublished work) that matched that observed with the native cyt bc1 in the presence of stigmatellin (Fig. 4). This resistance contrasts with the uninhibited native cyt bc1, which is readily proteolysed by thermolysin to release an 18-kDa Fe-S subunit fragment lacking its first 46 amino acid residues. Thus, the conformation of the +2Ala mutant either with or without stigmatellin resembles that of the native Fe-S subunit in the presence of stigmatellin.
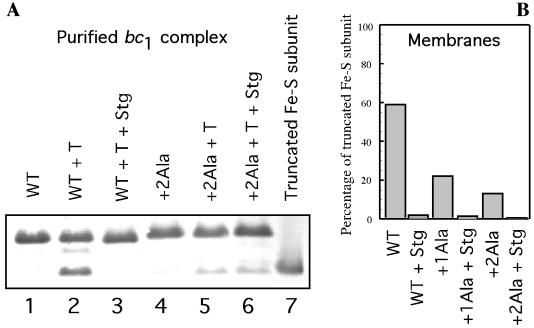
Figure 4.
Thermolysin-mediated proteolysis of purified or chromatophore membranes embedded R. capsulatus cyt bc1. (A) Purified cyt bc1 (0.6 nmol) from the wild type (lanes 1–3) or +2Ala mutant (lanes 4–6) was digested for 1 h with 0.2 nmol of thermolysin in the presence or absence of 20 μM stigmatellin. Aliquots were analyzed by immunoblotting with polyclonal antibodies against the Fe-S subunit of R. capsulatus. Lane 1, wild type (WT), nondigested; lane 2, wild type + thermolysin; lane 3, wild type + thermolysin + stigmatellin; lane 4, +2Ala mutant nondigested; lane 5, +2Ala mutant + thermolysin; lane 6, +2Ala mutant + thermolysin + stigmatellin; lane 7, 18-kDa fragment of the Fe-S subunit as a control (19). (B) Chromatophore membranes (650 μg) from the wild type, +1Ala mutant, or +2Ala mutant were digested for 1 h with 2 nmol thermolysin in the presence or absence of 20 μM stigmatellin. Aliquots were analyzed as described for A; the immunoblot was scanned, and the percentage of the 18-kDa fragment of the Fe-S subunit was calculated and plotted as a bar graph from left to right as wild type + thermolysin, wild type + thermolysin + stigmatellin, +1Ala mutant + thermolysin, +1Ala mutant + thermolysin + stigmatellin, +2Ala mutant + thermolysin, and +2Ala mutant + thermolysin + stigmatellin.
Movement of the Fe-S Subunit Extrinsic Domain Is Required for cyt bc1 Turnover but Not for QH2 Oxidation at the Qo Site.
Light-induced, single-turnover kinetics experiments monitoring cyt b reduction in the presence of antimycin and performed at 400 mV (a redox potential at which the [2Fe2S] is initially oxidized) revealed that transient cyt b reduction kinetics observed with the +2Ala mutant were similar to those seen with a wild-type strain (QH2 to cyt b about 50 s−1; data not shown). Therefore, the absence of the Fe-S subunit extrinsic domain movement does not prevent QH2 oxidation, even though it abolishes the turnover of the cyt bc1 by impeding the electron shuttling to cyt c1. This finding further highlights that the concerted electron transfer step is located between the cyt b and the Fe-S subunit of the cyt bc1.
Visualizing Electron Transfer from the [2Fe2S] Cluster to cyt c1 Heme with the +1Ala Mutant.
Remarkably, in the case of the +1Ala mutant, the cyt c reduction kinetics were now slow enough to lie between the wild type (too fast) and the +2Ala mutant (too slow), and hence they were measurable; in the presence and absence of myxothiazol, their half-times were 3 and 10 ms, respectively (Fig. 3C). Moreover, cyt c reduction kinetics could be rendered even more obvious by performing the experiments at pH 9.0. Under these conditions, the Em of the [2Fe2S] cluster (but not that of cyt c1 heme) decreases, thus shifting the redox equilibrium between these two cofactors to favor the reduction of cyt c1 (Fig. 3C, pH 9). Again, diminished cleavage by thermolysin of the +1Ala mutant cyt bc1 (Fig. 4), increased Q content trapped at the Qo site of the purified complex (EPR spectra not shown), and increased Em of the [2Fe2S] cluster (ΔEm = +60 mV; Table 1) indicated a more favored Qo position for the [2Fe2S] cluster domain.
It could be argued that the slower cyt c reduction kinetics observed in the alanine insertion mutants are due to the increased Em values of their [2Fe2S] clusters thermodynamically displacing the redox equilibrium between the [2Fe2S] cluster and cyt c1 heme. However, other mutants with increased Em values, such as the Fe-S subunit A46T mutant with Em7 =386 mV (Table 1), are still functional (21). Further, in the case of the +2Ala mutant, one would expect that in the uninhibited state, the cyt bc1 should still function despite this uphill reaction (25). Moreover, in the presence of myxothiazol, the Em differences vanish (E.D., and F.D., unpublished work). Therefore, such thermodynamic equilibrium cannot alone account for the impeded electron transfer observed in the alanine insertion mutants.
A second possibility could be an improper docking of the Fe-S subunit on cyt c1 as a possible consequence of the hinge-region mutations. However, in the mitochondrial cyt bc1 structure in which the Fe-S subunit extrinsic domain is in the c1 position, one of the histidine ligands of the [2Fe2S] cluster (His-161 in bovine numbering) is located close enough to one of the propionates of cyt c1 heme to form a hydrogen bond (8, 9). Electron-tunneling calculations indicate that electron transfer rates between these two cofactors should be extremely fast (≪1 μs; refs. 11 and 25). Thus, for the electron transfer between the [2Fe2S] cluster and cyt c1 heme to become rate limiting to the level observed in this study (more than several milliseconds), the [2Fe2S] cluster should dock in the alanine insertion mutants at least 5–8 Å away from its native position, which is highly unlikely.
On the other hand, the higher Em of the [2Fe2S] cluster, the higher Q content trapped in the Qo site on purification of the mutant cyt bc1, and the increased resistance to thermolysin-mediated conformation-sensitive cleavage combine with the slow cyt c kinetics to indicate a strongly favored Qo position for the extrinsic domain of the Fe-S subunit. Therefore, the slow electron transfer rates observed in these mutants are at least partially due to a hindered movement from the Q0 site to the cyt c1 position caused by the alanine residue insertions. Further support for this conclusion is provided by the successful isolation of a faster-growing revertant of the +1Ala mutant with a second site suppressor mutation located at position 286 (262 in bovine numbering) of the ef loop of R. capsulatus cyt b. This region represents the most conspicuous physical barrier that needs to be crossed during normal [2Fe2S] cluster domain movement (Fig. 1). Indeed, in this revertant, cyt c1 reduction kinetics in the presence of myxothiazol have a fast unresolved phase (<50 μs) exactly like that seen in the wild type (data not shown).
Implications of the [2Fe2S] Cluster Domain Movement on Energy Conversion.
The [2Fe2S] cluster domain motion on the 1- to 10-ms time scale in the +1Ala mutant may represent an engineering boundary at slower times, beyond which the movement begins to interfere significantly with the rate of electron transfer through the cyt bc1 and the rate of physiological growth (Fig. 5). Based on the motion resolved in the +1Ala mutant, it is clear that the movement in the native cyt bc1 must, as proposed, be faster than the 50-μs resolution of light-induced kinetics and hence be very much faster than the 1,700-s−1 kcat of the enzyme (12, 13). Thus, in the native enzyme, movement is required for catalytic activity but is neither rate-limiting nor essential for QH2 oxidation. Furthermore, our electron transfer simulations indicate that there may be another engineering boundary at faster times in the 1- to 10-μs range beyond which the cyt bc1 function may also become impaired (C.C.M., E.D., F.D., and P.L.D., unpublished work). These findings suggest that if the motion becomes too fast, then the system becomes vulnerable to short circuiting of electron transfer in the Qo site that inevitably leads to a less efficient energy conversion in the Q cycle mechanism (26).
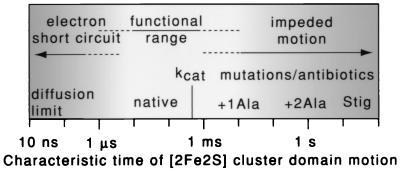
Figure 5.
Functional range of the [2Fe2S] cluster domain movement. The time scale of the [2Fe2S] cluster domain movement seems poised between two extremes of failure. Slowing the movement, as in the +1Ala mutant, to just less than the normal rate of Qo site catalysis kcat (characteristic turnover time of 600 μs) constrains the cyt bc1 turnover and compromises growth. Impeding the motion more drastically with stigmatellin, or as in the +2Ala mutant, blocks cyt bc1 catalysis and photosynthetic growth. Conversely, at faster times, simulations suggest that speeding the motion toward the physical diffusion limits (10 ns) may enable wasteful electron transfer and short circuit the redox-energy conversion mechanism.
Large amplitude domain motions therefore represent an effective design to control the flux of electrons inside protein complexes with branched electron transfer pathways. However, this unusual control exposes the cyt bc1 to antibiotics designed to act, not as usual by displacing the substrate from the catalytic sites, but instead on the domain motion. This type of control might be achieved by impeding the helix-random coil conformational change in the [2Fe2S] hinge region, which, based on various structures (8–10) and mutants (24, 27), is required for the movement. The intraprotein electron shuttle motion of the Fe-S subunit demonstrated herein can now be explored by using “slow” mutants to analyze how this movement is controlled at the molecular level.
Acknowledgments
We would like to thank B. R. Gibney and R. E. Sharp for assistance with the EPR and flash kinetics spectroscopy. This work was supported by National Institutes of Health Grants GM 38237 to F.D. and GM 27309 to P.L.D.
Abbreviations
- cyt
cytochrome
- Q
ubiquinone
- QH2
ubihydroquinone
- EPR
electron paramagnetic resonance
References
- 1.Finer J T, Simmons R M, Spudich J A. Nature (London) 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 2.De Rosier D J. Cell. 1998;93:17–20. [Google Scholar]
- 3.Sabbert D, Engelbrecht S, Junge W. Nature (London) 1996;381:623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- 4.Sambongi Y, Iko Y, Tanabe M, Omote H, Iwamoto-Kihara A, Ueda I, Yanagida T, Wada Y, Futai M. Science. 1999;286:1722–1724. doi: 10.1126/science.286.5445.1722. [DOI] [PubMed] [Google Scholar]
- 5.Gray K A, Daldal F. In: Anoxygenic Photosynthetic Bacteria. Blankenship R E, Madigan M T, Bauer C, editors. Dordrecht, The Netherlands: Kluwer; 1995. pp. 747–774. [Google Scholar]
- 6.Cramer W, Soriano G, Ponomarev M, Huang D, Zhang H, Martinez S, Smith J. Annu Rev Plant Physiol. 1996;47:477–508. doi: 10.1146/annurev.arplant.47.1.477. [DOI] [PubMed] [Google Scholar]
- 7.Xia D, Yu C-A, Kim H, Xia J-Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, Huang L, Shulmeister V M, Chi Y-I, Kim K K, Hung L-W, Crofts A R, Berry E A, Kim S-H. Nature (London) 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 9.Iwata S, Lee J W, Okada K, Lee J K, Iwata M, Rasmussen B, Link T A, Ramaswamy S, Jap B K. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 10.Kim H, Xia D, Yu C-A, Xia J-Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Proc Natl Acad Sci USA. 1998;95:8026–8033. doi: 10.1073/pnas.95.14.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moser C C, Keske J M, Warncke K, Farid R S, Dutton P L. Nature (London) 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 12.Crofts A R, Wang Z. Photosynth Res. 1989;22:69–87. doi: 10.1007/BF00114768. [DOI] [PubMed] [Google Scholar]
- 13.Ding H, Moser C C, Robertson D, Tokito M, Daldal F, Dutton P L. Biochemistry. 1995;34:15979–15996. doi: 10.1021/bi00049a012. [DOI] [PubMed] [Google Scholar]
- 14.Ding H, Robertson D, Daldal F, Dutton P L. Biochemistry. 1992;31:3144–3158. doi: 10.1021/bi00127a015. [DOI] [PubMed] [Google Scholar]
- 15.Ohnishi T, Brandt U, von Jagow G. Eur J Biochem. 1988;176:385–389. doi: 10.1111/j.1432-1033.1988.tb14293.x. [DOI] [PubMed] [Google Scholar]
- 16.Valkova-Valchanova M, Saribas A S, Gibney B R, Dutton P L, Daldal F. Biochemistry. 1998;37:16242–16251. doi: 10.1021/bi981651z. [DOI] [PubMed] [Google Scholar]
- 17.Atta-Asafo-Adjei E, Daldal F. Proc Natl Acad Sci USA. 1991;88:492–496. doi: 10.1073/pnas.88.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson D E, Ding H, Chelminski P R, Slaughter C, Hsu J, Moomaw C, Tokito M, Daldal F, Dutton P L. Biochemistry. 1993;32:1310–1317. doi: 10.1021/bi00056a016. [DOI] [PubMed] [Google Scholar]
- 19.Saribas A S, Valkova-Valchanova M, Tokito M K, Zhang Z, Berry E A, Daldal F. Biochemistry. 1998;37:8105–8114. doi: 10.1021/bi973146s. [DOI] [PubMed] [Google Scholar]
- 20.Dutton P L. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 21.Brasseur G, Sled V, Liebl U, Ohnishi T, Daldal F. Biochemistry. 1997;36:11685–11696. doi: 10.1021/bi970777d. [DOI] [PubMed] [Google Scholar]
- 22.Gray K, Dutton P L, Daldal F. Biochemistry. 1994;33:723–733. doi: 10.1021/bi00169a014. [DOI] [PubMed] [Google Scholar]
- 23.Saribas A S, Ding H, Dutton P L, Daldal F. Biochemistry. 1995;34:16004–16012. doi: 10.1021/bi00049a014. [DOI] [PubMed] [Google Scholar]
- 24.Darrouzet E, Valkova-Valchanova M, Ohnishi T, Daldal F. J Bioenerg Biomembr. 1999;31:275–288. doi: 10.1023/a:1005428014548. [DOI] [PubMed] [Google Scholar]
- 25.Page C C, Moser C C, Chen X, Dutton P L. Nature (London) 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell P. FEBS Lett. 1975;59:137–139. doi: 10.1016/0014-5793(75)80359-0. [DOI] [PubMed] [Google Scholar]
- 27.Tian H, White S, Yu L, Yu C-A. J Biol Chem. 1999;274:7146–7152. doi: 10.1074/jbc.274.11.7146. [DOI] [PubMed] [Google Scholar]