Abstract
Mutations in palmitoyl-protein thioesterase 1 (PPT1), a lysosomal enzyme that removes fatty acyl groups from cysteine residues in modified proteins, cause the fatal inherited neurodegenerative disorder infantile neuronal ceroid lipofuscinosis. The accumulation of undigested substrates leads to the formation of neuronal storage bodies that are associated with the clinical symptoms. Less severe forms of PPT1 deficiency have been found recently that are caused by a distinct set of PPT1 mutations, some of which retain a small amount of thioesterase activity. We have determined the crystal structure of PPT1 with and without bound palmitate by using multiwavelength anomalous diffraction phasing. The structure reveals an α/β-hydrolase fold with a catalytic triad composed of Ser115-His289-Asp233 and provides insights into the structural basis for the phenotypes associated with PPT1 mutations.
Infantile neuronal ceroid lipofuscinosis (INCL) is the most severe form of the neuronal ceroid lipofuscinoses (NCLs), a family of recessively inherited childhood neurodegenerative disorders. Infants with INCL are normal until 1–2 years of age, when they develop retinal blindness, ataxia, and seizures. Affected children typically show a flat electroencephalogram by age 3, and death occurs most often by ages 8–11 (1).
Mutations in a gene encoding the novel hydrolytic enzyme palmitoyl protein thioesterase 1 (PPT1) are the underlying cause of INCL (2). The PPT1 gene was identified as a candidate for the disorder when it was mapped to chromosome 1p32, a region known to contain the defective gene in a group of Finnish patients (3). Over 20 different mutations in the PPT1 gene have been identified in NCL patients worldwide (4).
PPT1 is a lysosomal enzyme (5) that plays a major role in the degradation of lipid-modified proteins, removing fatty acids (usually palmitate) from cysteine residues (6). The enzyme was originally isolated by virtue of its ability to remove palmitate from the proto-oncoprotein H-Ras (7, 8), but it has a broad substrate specificity, cleaving fatty acyl chains of lengths 14–18 carbons from cysteine residues in proteins and peptides as well as from palmitoyl-CoA (8) and S-palmitoyl thioglucoside (9). Palmitoylation is an important protein modification that facilitates membrane localization and protein-protein interactions (10). A cytosolic acyl-protein thioesterase has been described that may play a role in agonist-stimulated turnover of palmitate on proteins (11); however, the role of PPT1 is clearly distinct from acyl-protein thioesterase and appears to be confined to the lysosomal degradation of protein-derived fatty acyl thioesters.
Over two dozen distinct enzymatic reactions are catalyzed by thioesterases, but three-dimensional structures for only two thioesterases have been determined previously. The Vibrio harveyi myristoyl-acyl carrier protein thioesterase (ACTE) (12), an enzyme involved in bioluminescence, contains the α/β hydrolase fold characteristic of microbial lipases. The second, 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. strain CBS-3 (13), an enzyme of interest in bioremediation, is unrelated in both structure and mechanism to the α/β hydrolases. PPT1 represents the only eukaryotic thioesterase to be examined at atomic resolution.
We determined the crystal structure of bovine PPT1 by using the multiwavelength anomalous diffraction phasing method on a crystal of selenomethionine (SeMet) labeled PPT1. Bovine PPT1 is 95% identical to human PPT1 at the amino acid level (14). The structure has been refined to 2.25 Å resolution. We have also determined the structure of a covalent acyl-enzyme complex between PPT1 and palmitate, which has been refined to 2.5 Å resolution. The structure of the acyl-enzyme intermediate confirmed the identity of Ser115 as the nucleophile and identified the hydrophobic groove that accommodates the fatty acid chain.
Materials and Methods
Protein Expression and Purification.
Native and SeMet-labeled bovine PPT1 for the structure determination were overexpressed in insect cells as reported (7, 15). PPT1 was secreted into the cell medium. One liter of crude medium was adjusted to 20 mM Hepes (pH 7.0) and 500 mM NaCl (Buffer A) and was applied to a 20-ml PhenylSepharose 6 Fast Flow (Low-Sub) column (Pharmacia) equilibrated with Buffer A. The column was washed sequentially with Buffer A; 20 mM Hepes (pH 7.0) and 150 mM NaCl (Buffer B); 5 mM Hepes (pH 7.0); and a gradient from 0 to 75% ethylene glycol in 5 mM Hepes (pH 7.0). This protocol was sufficient to remove virtually all other proteins from the column, but PPT1 remained tightly bound to the column and required an elevated pH for elution. PPT1 was eluted with 5 mM Hepes (pH 7.9) and 75% ethylene glycol. Because PPT1 is unstable under basic conditions, it was eluted into tubes containing 1 M Hepes (pH 7.0) to immediately lower the pH. We then dialyzed the PPT1 against Buffer B to remove the ethylene glycol and concentrated it to 4 ml. The sample was further purified by gel filtration in Buffer B on a Sephacryl S-100 26/60. The purified PPT1 was concentrated to 12 mg/ml in Buffer B. The average yield of purified PPT1 was approximately 10 mg per liter of cell culture medium.
Crystallization and Structure Determination.
PPT1 crystals grew from sitting drop vapor diffusion experiments containing 3 μl of 12 mg/ml PPT1 in Buffer B and 2 μl of reservoir solution equilibrated against a 500-μl reservoir containing 55% polypropylene glycol 400 (Fluka) and 100 mM Bis-Tris (pH 6.5). Crystals of SeMet PPT1 were obtained under identical conditions. Crystals of the PPT1/palmitate complex were obtained by using the above conditions with 12 mg/ml PPT1 in Buffer B with 2 mg/ml palmitoyl-CoA. For data collection, crystals were harvested from the drop in nylon loops and were frozen in liquid nitrogen.
Data collection and refinement statistics appear in Table 1. Multiwavelength anomalous diffraction data at three wavelengths were collected by using the inverse beam technique on a single SeMet PPT crystal using an ADSC Quantum-4 CCD detector (16) on the F2 beamline at the Cornell High Energy Synchrotron Source (CHESS). The data were indexed and integrated by using mosflm (17) and were scaled and truncated with scala and truncate from the Collaborative Computational Project 4 package (18). The selenium atoms were located by using solve (19), and heavy atom refinement and phasing was accomplished with sharp (20). After density modification with solomon (21), the figure of merit for all reflections was 0.91. The experimental electron density was readily interpretable (Fig. 3A), and the model was built by using o (22). The model was refined against a 2.25 Å native PPT1 dataset collected at the CHESS A1 beamline. The native dataset was processed with the hkl suite (23). Refinement was carried out by using cns (24) with manual rebuilding in o. Diffraction data on the complex were collected at the BioCARS 14BM-D beamline at the Advanced Photon Source and were processed in the same manner as the SeMet data. Density for the palmitate was clearly visible in difference Fourier maps. The palmitate was added to the model by using o and was refined as above.
Table 1.
Data collection, phasing, and refinement statistics
| Native | SeMet λ1 | SeMet λ2 | SeMet λ3 | Complex | |
|---|---|---|---|---|---|
| Beamline | CHESS A1 | CHESS F2 | CHESS F2 | CHESS F2 | APS 14BM-D |
| Unit cell, Å | a = b = 69.35 | a = b = 68.11 | a = b = 68.11 | a = b = 68.11 | a = b = 67.52 |
| c = 128.39 | c = 127.31 | c = 127.31 | c = 127.31 | c = 128.92 | |
| Wavelength, Å | 0.9100 | 0.9791 | 0.9789 | 0.9364 | 1.0088 |
| Resolution, Å | 20.9–2.25 | 42.4–3.0 | 42.4–3.0 | 42.4–3.0 | 41.2–2.5 |
| Observed reflections | 109,210 | 165,466 | 138,024 | 119,571 | 113,623 |
| Unique reflections | 14,788 | 7,148 | 7,144 | 6,816 | 10,620 |
| Completeness (%) | 94.8 (91.4) | 100.0 (100.0) | 100.0 (100.0) | 99.8 (99.8) | 97.4 (97.8) |
| Rsym | 0.055 (0.164) | 0.072 (0.324) | 0.078 (0.381) | 0.078 (0.344) | 0.148 (0.319) |
| Ranomalous | 0.055 | 0.055 | 0.054 | ||
| Rdispersive | 0.021 (λ2–λ1) | 0.033 (λ3–λ2) | 0.039 (λ3–λ1) | ||
| Rworking (Rfree) | 0.241 (0.297) | 0.226 (0.270) | |||
| Number of nonhydrogen atoms | |||||
| Protein | 2210 | 2210 | |||
| Carbohydrate | 55 | 55 | |||
| Palmitate | 17 | ||||
| Water | 53 | 36 | |||
| RMS deviations from ideal values | |||||
| Bond lengths, Å | 0.008 | 0.008 | |||
| Bond angles, degrees | 1.3 | 1.4 |
Figure 3.
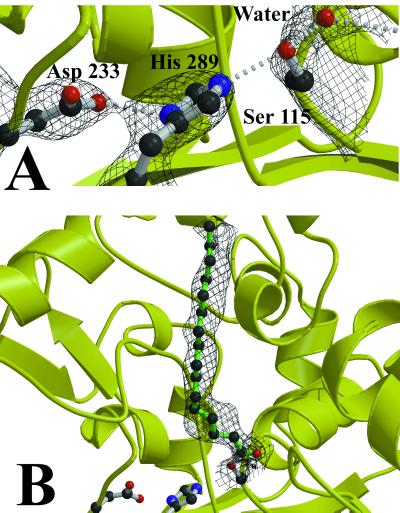
Active site of PPT1. (A) The active site of native PPT1. A water molecule occupies the oxyanion hole, hydrogen-bonded to Ser115, Met41, and Gln116. The solvent-flattened multiwavelength anomalous diffraction electron density with experimental phases is superimposed on the model, contoured at 1.4σ. (B) The active site of the PPT1/palmitate complex. A simulated annealing Fo-Fc map, generated by using cns (24) with Ser115 and palmitate omitted from the calculation, is shown contoured at 1.5σ.
Site-Directed Mutagenesis.
The plasmid pCMV5-hPPT was constructed by PCR amplification of the entire coding region of the human PPT cDNA using pSV-hPPT (2) with insertion of the amplified product into the EcoRI-HindIII sites of pCMV5 (25). Site-directed mutations were introduced by using a commercial method (Chameleon, Stratagene). An oligonucleotide primer containing a ScaI to MluI mutation within the ampicillin resistance gene of pCMV5 was used to select mutagenized plasmids. Double- and triple-site asparagine-linked glycosylation mutants were constructed by iterative rounds of subcloning and mutagenesis. All plasmid coding regions were verified by DNA sequencing.
Effect of Site-Directed Mutations on PPT Activity.
Simian COS-1 cells were maintained as described (6). Cells growing in 60-mm plates were transiently transfected with 2 μg of DNA and 10 μl of FuGENE 6 reagent (Boehringer Mannheim). Cells were harvested at 65–72 h posttransfection into buffer containing 50 mM Tris⋅HCl (pH 7.0), 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 2 μg/ml of leupeptin, 1 μg/ml of pepstatin, and 2 mM PMSF and were homogenized by sonication. The homogenates were centrifuged at 12,000 × g for 10 min, and supernatants were assayed for PPT activity by monitoring the release of [3H]palmitate from [3H]palmitoyl-CoA (26). Assays were normalized for PPT1 protein expression by immunoblotting and scanning densitometry, and were corrected for endogenous background PPT activity by using cells transfected in parallel with a plasmid vector containing no cDNA insert. The background averaged 13.5 ± 1.4% of the wild-type level for each experiment. The expression level of mutant PPT1 was similar (50–100%) to that of wild-type controls, with the exception of N197Q/N212Q, which showed expression at 10% of the wild-type level (data not shown).
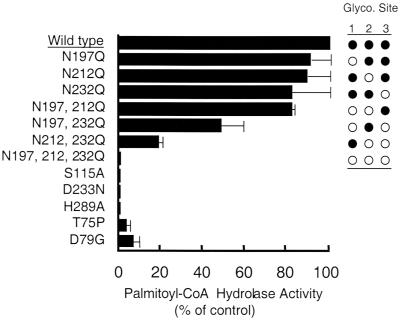
Asparagine-to-glutamine substitutions were made at each of the three glycosylation sites (Fig. 1). Elimination of glycosylation at each mutated site was verified by comparison of the migration of the expressed protein with the wild-type by SDS/PAGE and immunoblotting (data not shown).
Figure 1.
Palmitoyl-CoA hydrolase activity in COS cell lysates expressing wild-type and mutant human PPT1s. Activity is expressed as a percentage of wild-type activity over control cells transfected with vector alone. Each value represents the mean ± SE from three or more independent experiments. As confirmed by the crystal structure, Ser115, Asp233, and His289 are essential for activity and comprise the catalytic triad of PPT1. To assess the effect of glycosylation site mutations on PPT1 activity, the three asparagine glycosylation sites in PPT1 were mutated singly or in combination as shown. Filled circles indicate glycosylation site occupancy. Two mutations associated with juvenile onset NCL, Thr75Pro and Asp79Gly, are shown to exhibit detectable residual PPT activity.
Results and Discussion
Structure Determination of PPT1.
Although crystals of recombinant PPT1 were obtained readily, they showed large variances in diffraction limit and unit cell parameters. Very high merging R-factors from multiple crystals indicated that native crystals were non-isomorphous to each other, making the success of isomorphous replacement phasing unlikely. After a systematic study of growth conditions, we were able to express SeMet-labeled PPT1 in the baculovirus system with sufficient incorporation of SeMet to solve the structure by multiwavelength anomalous diffraction phasing (15).
Overall Architecture of PPT1.
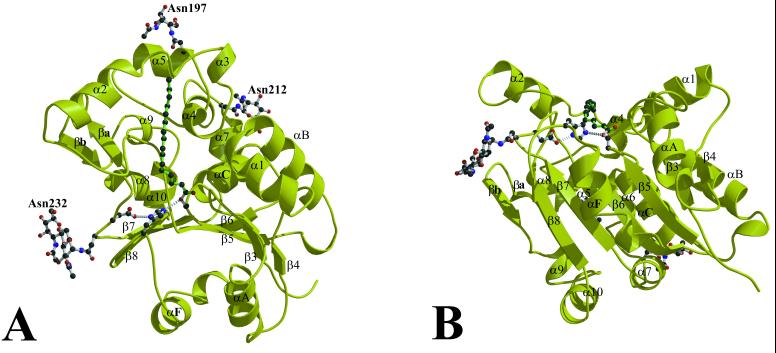
The final refined model contains residues 28–306, which corresponds to the entire mature PPT1 polypeptide after cleavage of the 27-residue signal peptide. PPT1 adopts an α/β-hydrolase fold, with a central six-stranded parallel β-sheet corresponding to strands β3–β8 in the α/β hydrolase nomenclature (27) (Fig. 2). Topologically, PPT1 is similar to the haloperoxidase subfamily (which includes ACTE) and the dienlactone hydrolase subfamily, although it lacks the antiparallel β strands β1 and β2. Like these two subfamilies, PPT1 has an insertion between β4 and αB. In PPT1, this insertion contains a helix (α1) that is roughly perpendicular to the direction of the β-sheet (Fig. 2). There is also a large insertion between β6 and β7 (residues 140–223) which forms a second domain that encompasses most of the fatty acid binding site. This region includes six helices (α2-α7). None of the helices in this insertion correspond to αD in the canonical α/β-hydrolase fold, which is antiparallel to the β sheet. PPT1 is also missing the canonical antiparallel helix αE, having instead another insertion between β7 and β8 containing several helices and a loop connecting two short antiparallel β-strands (βa and βb).
Figure 2.
The crystal structure of PPT1 complexed with palmitate. β3–β8 and αA, αB, αC, and αF are elements of the canonical α/β hydrolase fold and are labeled according to the nomenclature of Ollis (27). The palmitate, the catalytic triad, and the glycosylated asparagines are indicated. B is a rotation by 90° from A. Figs. 2, 3, and 5 were prepared by using bobscript (28), molscript (29), and raster3d (30).
Catalytic Triad of PPT1.
PPT1 has a catalytic triad composed of Ser115, His289, and Asp233 (Fig. 3A). The nucleophilic Ser115 of PPT1 is located on a loop between β5 and αC with the unusual geometry (φ = 60.0°, ψ = −128.7°) characteristic of the nucleophile elbow of α/β hydrolases. Although we had previously used site-directed mutagenesis to identify Ser115 and His289 as critical residues for activity (Fig. 1), the identity of the third member of the catalytic triad could not be ascertained before the completion of the structure determination, as several candidates were suggested by the mutagenesis data. Asp233 was a surprising finding in light of the fact that it is present in the recognition site for N-linked glycosylation at Asn232.
Glycosylated Asparagines of PPT1.
There are three potential glycosylation sites in the sequence of the mature PPT1 polypeptide. The electron density is sufficient to model one N-acetyl glucosamine residue attached to Asn197 and Asn212, and two N-acetyl glucosamine residues joined in a β1–4 linkage attached to Asn232.
Mutation of the glycosylated asparagines to glutamine leads to a reduction in thioesterase activity of the mutant protein (Fig. 1). Omitting any one of the three glycosylation sites produces protein that has activity comparable to the wild type. Double mutants show a reduction in activity that depends on which sites are blocked; those containing Asn232Gln mutations are less active than the Asn197Gln/Asn212Gln double mutant. This is consistent with Asn232's proximity to the catalytic Asp233. The triple mutant has no detectable thioesterase activity. We suspect that this inactive nonglycosylated protein is improperly folded and probably insoluble, in view of our inability to overexpress soluble PPT1 in Escherichia coli, which is incapable of adding N-linked glycosylation.
Structure of the PPT1/Palmitate Complex.
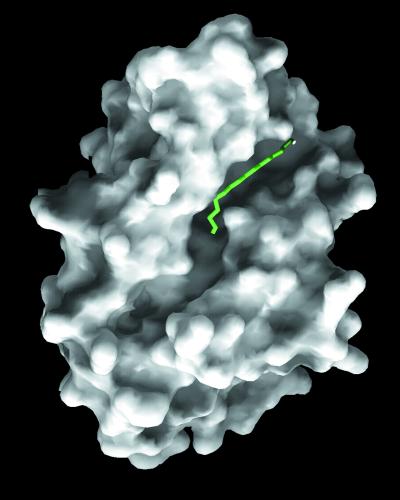
The structure of the covalent acyl-enzyme intermediate illuminates the fatty acid binding site, as shown in Figs. 3B and 4. The palmitate binds in a mostly extended conformation in a hydrophobic groove in the second domain of PPT1. The C4-C5 bond in the palmitate adopts a gauche conformation, producing a bend in the chain. The fact that the palmitate has to bend to fit into the binding pocket may suggest that the pocket is designed to bind an unsaturated fatty acid with a cis-double bond between C4 and C5 (although such fatty acids are rarely found in nature).
Figure 4.
The palmitate binding groove. The molecular surface was calculated by using grasp (31). The palmitate (green stick model) occupies a deep, narrow groove on the hydrophobic face of the protein.
Nucleophilic attack by Ser115 on the carbonyl of palmitate generates a tetrahedral intermediate, and the main-chain amide nitrogens of Met41 and Gln116 can act as hydrogen bond donors to stabilize the intermediate oxyanion. In the uncomplexed structure, a water molecule hydrogen bonds to these two nitrogens and to Ser115 (Fig. 3A). In the complex, Met41 and Gln116 are within hydrogen bonding distance of the palmitate carbonyl oxygen (Fig. 3B).
The residues lining the hydrophobic binding groove come from several secondary structural elements. The top of the groove is formed by residues from α2 and α3 in the second domain: Leu167, Asn 168, Ala171, Tyr172, and Ile176. The loop connecting β6 to the beginning of the second domain contributes Val 146, Leu 149, and Pro150. Helix α4 and the loop immediately after it contribute Leu180, Gln182, Ala183, Tyr185, and Trp186 to the binding groove. Finally, several residues near the active site form the rest of the groove: Ile235, Val236, Gln116, Gly40, and Met41.
Gly40, Met41, Gln116, Gln142, Val146, Pro150, Ala171, Tyr172, Gln182, Ala183, Tyr185, Trp186, and Val236 are invariant in all PPT1 isoforms from Caenorhabditis elegans to humans (Gln116, Val146, Pro150, Tyr172, and Tyr185 are also conserved in Arabidopsis) (32). The other residues lining the binding site show only conservative substitutions in the known PPT1s.
There are no significant differences observed in the crystal structures between the uncomplexed and complexed forms of PPT1. In particular, there is no evidence for a movable lid of the type that regulates interfacial activation of certain lipases (33). No evidence of interfacial activation has been found when palmitoyl-CoA has been used as a substrate in kinetic studies.
PPT1 shows structural and mechanistic similarity to ACTE despite a lack of sequence homology. The two thioesterases share the core six-stranded β-sheet, although ACTE also contains the antiparallel strands β1 and β2 at the N-terminal end, as well as a longer C terminus, which contains an additional α-helix and β-strand. The insertion between β6 and β7 that forms the palmitate binding domain corresponds topologically to the smaller “cap” subdomain predicted form the myristoyl-binding pocket in ACTE (12), but it is structurally quite different. PPT1 and ACTE have a common catalytic triad and acyl transfer mechanism, but differ in specificity. Notably, ACTE prefers 14-carbon acyl esters and thioesters (34) whereas PPT1 acts on a broader range of fatty acyl chain lengths, but only hydrolyzes thioesters and not esters (7).
Molecular Basis for NCL.
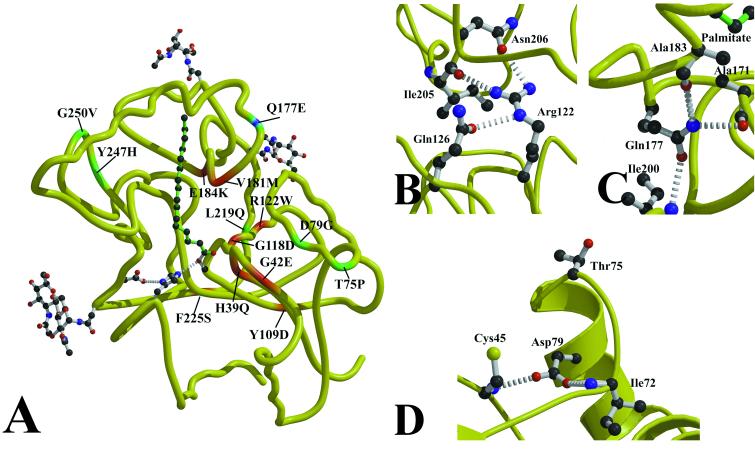
A number of clinically significant mutations in PPT1 have been identified (Fig. 5A) (4, 32, 35). The common Finnish variant of INCL is caused by a single missense mutation (Arg122Trp) in PPT1 that leads to a misfolded enzyme that is trapped in the endoplasmic reticulum (2). Lymphoblasts derived from subjects with this mutation have no detectable PPT1 activity (36). Arg122 is located on αC, immediately after the nucleophile elbow. The sidechain of Arg122 extends toward α6 and acts to control the spacing between those two helices (Fig. 5B). The terminal guanidinium of Arg122 is hydrogen-bonded to the main-chain carbonyl of Ile205 and Oδ of Asn206 on α6. There is also a third hydrogen bond between Nɛ of Arg122 and Oɛ of Gln126. Mutation of Arg122 to Trp means not only a loss of those three hydrogen bonds but also a steric and polarity mismatch with the surrounding residues. These steric clashes would disrupt the folding of PPT1's core by pushing αC away from α6, β7, or the loop connecting β6 to the palmitate-binding domain. The buried polar pocket containing Arg122 appears to be similar to ones found in some fungal lipases (37), which are also located near the active site and in the C-terminal half of the β-sheet.
Figure 5.
NCL mutations in PPT1. (A) Sites of clinical NCL mutations in PPT1 are mapped onto the peptide backbone. INCL mutations are displayed in red, a mutation causing LINCL symptoms is in blue, and JNCL mutations are in green. (B) The most common INCL mutation (Arg122Trp) leads to the loss of three hydrogen bonds and a steric and polarity mismatch with the surrounding residues, resulting in misfolded protein. (C) The Gln177Glu mutation is predicted to cause the loss of hydrogen bonds to Ala171 and Ala183, two residues that contact palmitate. This mutation results in a less severe phenotype that is clinically indistinguishable from classical LINCL. (D) Two mutations on α1 lead to JNCL. Trp75Pro may alter the beginning of α1 due to the conformational restraints on Pro, and Asp79Glu loses hydrogen bonds to Cys45 and Ile72.
A number of other PPT1 mutations have been detected in INCL patients (4, 32, 35). Many of these are truncations caused by nonsense or deletion mutations, and these presumably lead to inactive PPT1 due to the proximity of the catalytic histidine to the C terminus. A number of missense mutations have also been characterized, and these can be interpreted in the context of the crystal structure.
Several INCL mutations are predicted to cause unfavorable steric, polar, and/or electrostatic interactions that could perturb the geometry of the nucleophile elbow, which is responsible for the proper location and orientation of Ser115. Catalytic efficiency should be highly sensitive to the geometry of Ser115 because it must be activated by His289 and in position to attack the substrate.
The INCL mutation Gly118Asp is immediately adjacent to the nucleophile elbow region. The Asp sidechain of this mutant is predicted to extend into a hydrophobic pocket containing sidechains from Phe225, Val136, and Leu123, causing steric and polarity clashes. Likewise, Tyr109Asp would introduce a charged sidechain into a hydrophobic cluster made up of Val36, Leu99, Leu105, Val124, and Met133. Because Tyr109 is located at the opposite end of β4 from the nucleophile, it is possible that a very small movement of the backbone due to unfavorable interactions between the Asp sidechain and neighboring residues could lead to a significant displacement of Ser115 due to leverage along the β-strand.
Another severe mutation, Phe225Ser (W. Yi and S.L.H., unpublished data), is located in a hydrophobic pocket consisting of sidechains from β6, β7, and β8, which is adjacent to the polar cluster containing the Arg122 sidechain. The introduction of a polar side chain into this pocket could disrupt the hydrophobic sidechain packing and adversely affect the folding of the core β-sheet. Additionally, the Ser hydroxyl may be able to interact with the Arg122 sidechain, and thereby disrupt the hydrogen bonding network between αC and α6 as in the Arg122Trp mutation.
There are two INCL mutations near Met 41 (Gly42Gln and His39Asp), a residue that plays two important roles in the active site of PPT1. The amide stabilizes the oxyanion intermediate, and the side chain contacts the palmitate. These mutations could disrupt the active site by perturbing the position of Met41, thus decreasing the oxyanion stabilization or altering the binding pocket. Gly42 is located on a β-turn with φ/ψ angles of 82.9° and 1.4°, respectively. It is unlikely that the Gly42Glu mutant could adopt this conformation, and a more energetically favorable conformation would alter the loop between β3 and αA, where Met41 is located. The other INCL mutation in this region, His39Gln, would push β3 away from the loop connecting it with αA by replacing His39 with a larger glutamine sidechain and disrupting a hydrogen bond between the His39 imidazole nitrogen and the main-chain carbonyl of Asp43.
Two INCL mutations (Val181Met and Glu184Lys) are located on α4 in the palmitate-binding domain. Several residues on this helix make hydrophobic contacts with the palmitate. The Val181Met mutation would push α4 and α1 away from each other because of the larger size of the Met sidechain. Likewise, the larger size and positive charge of the Lys side chain compared with Glu would push α4 away from αB in the Glu184Lys mutant. Either mutation probably shifts α4 in a manner that disrupts the palmitate-binding pocket.
Classical late-infantile NCL (LINCL) and juvenile NCL (JNCL), two other well-studied forms of NCL, are caused by mutations in other genes (CLN2 and CLN3, respectively). CLN2 codes for a lysosomal pepstatin-insensitive acid protease that is deficient in LINCL patients (38); the gene product of CLN3 is a lysosomal membrane protein of unknown function (39–41). LINCL and JNCL differ from INCL in the age of onset and severity of symptoms, and by differences in morphology of the lysosomal storage material (granular osmiophilic deposits in INCL, curvilinear inclusions in LINCL, and fingerprint inclusions in JNCL). Recently, mutations in PPT1 that produce symptoms clinically indistinguishable from LINCL and JNCL have been identified (4, 42). The storage bodies from these patients consist of the granular osmiophilic deposits characteristic of PPT1 deficiency.
One PPT1 mutation described in LINCL patients (35) is the small change of Gln177 to Glu. Gln177 participates in three structure-determining interactions, one as a hydrogen bond acceptor (from Ile200) and two as a hydrogen bond donor (to Ala171 and Ala 183) (Fig. 5C). In the Gln177Glu mutant, there will no longer be an Nɛ to act as an H-bond donor, so presumably there will be some reorganization of the conformation of the 177 side chain and surrounding residues and loss of the hydrogen bonds to Ala183 and Ala171. Because Ala171 and Ala183 both make hydrophobic contact with the palmitate, this could lead to a reduction in activity and the consequent clinical phenotype. These are presumably less dramatic changes to the binding pocket than those caused by Val181Met and Glu184Lys, which correlates with the severity of clinical symptoms.
In contrast to the INCL and LINCL mutations, which for the most part seem to perturb the active site and binding pocket geometry or disrupt the folding of the PPT1 core, the mutations associated with JNCL are located away from the active site and are predicted to cause less dramatic changes to the structure. Interestingly, some of these mutant proteins have been shown to retain a low level of PPT1 activity (Fig. 1), which could explain the milder phenotype (4).
The mutations associated with JNCL are found in two locations. Thr75Pro and Asp79Gly are located on α1, and Tyr247His and Gly250Val are located in the anti-parallel β-strand region between β7 and α8. Thr75 is located at the start of helix α1; mutation to a proline may perturb the geometry of the helix (Fig. 5D). The Asp79 sidechain is hydrogen-bonded to Cys45 and Ile72 (Fig. 5D). Loss of these hydrogen bonds on mutation to Gly could lead to increased flexibility in this region. On the opposite site of the molecule, Gly250 is located in a β-turn between βa and βb and has a positive ψ angle, so mutation to any other amino acid would not preserve this antiparallel β-motif. Tyr247 is located on βa, where it is hydrogen-bonded to Asp237 and through that interaction holds the antiparallel β-motif to the loop connecting it to the main β-sheet of the core domain.
In summary, the structural analysis of PPT1 in the context of known mutations is consistent with the idea that mutations that affect catalysis or substrate binding or disrupt proper folding of the core result in inactive enzymes and lead to a severe clinical phenotype. Other mutations associated with a less severe clinical course can, in some cases, be shown to retain some residual thioesterase activity, and all of these less severe mutations are predicted to make small, local changes in regions of the structure that are remote from the catalytic triad and palmitate binding site.
Acknowledgments
S.L.H., J.-Y.L., and A.K.D. thank Ahmad Gaben for technical assistance. This work was supported in part by a National Institutes of Health Training Grant in Molecular Biophysics (to J.J.B.), National Institutes of Health Grant CA59021 (to J.C.), New York State Sea Grant R/XBP-6 (to J.C.), and National Institutes of Health grants NS 35323 and NS36867 (to S.L.H.) and the Robert A. Welch Foundation (S.L.H.). This work is based in part on research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under award DMR-9311772, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award RR-01646 from the National Institutes of Health. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Energy Research, under Contract No. W-31-109-Eng-38.
Abbreviations
- PPT1
palmitoyl protein thioesterase 1
- INCL
infantile neuronal ceroid lipofuscinosis
- LINCL
late infantile neuronal ceroid lipofuscinosis
- JNCL
juvenile neuronal ceroid lipofuscinosis
- SeMet
selenomethionine
- ACTE
myristoyl-ACP thioesterase
- CHESS
Cornell High Energy Synchrotron Source
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1EI9 and 1EH5).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080508097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080508097
References
- 1.Santavuori P, Haltia M, Rapola J. Dev Med Child Neurol. 1974;16:644–653. doi: 10.1111/j.1469-8749.1974.tb04183.x. [DOI] [PubMed] [Google Scholar]
- 2.Vesa J, Hellsten E, Verkruyse L A, Camp L A, Rapola J, Santavuori P, Hofmann S L, Peltonen L. Nature (London) 1995;376:584–587. doi: 10.1038/376584a0. [DOI] [PubMed] [Google Scholar]
- 3.Hellsten E, Vesa J, Speer M C, Makela T P, Jarvela I, Alitalo K, Ott J, Peltonen L. Genomics. 1993;16:720–725. doi: 10.1006/geno.1993.1253. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann S L, Das A K, Yi W, Lu J-Y, Wisniewski K E. Mol Genet Metab. 1999;66:234–239. doi: 10.1006/mgme.1999.2803. [DOI] [PubMed] [Google Scholar]
- 5.Verkruyse L A, Hofmann S L. J Biol Chem. 1996;271:15831–15836. doi: 10.1074/jbc.271.26.15831. [DOI] [PubMed] [Google Scholar]
- 6.Lu J-Y, Verkruyse L A, Hofmann S L. Proc Natl Acad Sci USA. 1996;93:10046–10050. doi: 10.1073/pnas.93.19.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camp L A, Hofmann S L. J Biol Chem. 1993;268:22566–22574. [PubMed] [Google Scholar]
- 8.Camp L A, Verkruyse L A, Afendis S J, Slaughter C A, Hofmann S L. J Biol Chem. 1994;269:23212–23219. [PubMed] [Google Scholar]
- 9.van Diggelen O P, Keulemans J L, Winchester B, Hofman I L, Vanhanen S L, Santavuori P, Voznyi Y V. Mol Genet Metab. 1999;66:240–244. doi: 10.1006/mgme.1999.2809. [DOI] [PubMed] [Google Scholar]
- 10.Bizzozero O A. Neuropediatrics. 1997;28:23–26. doi: 10.1055/s-2007-973660. [DOI] [PubMed] [Google Scholar]
- 11.Duncan J, Gilman A. J Biol Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- 12.Lawson D, Derewenda U, Serre L, Ferri S, Szittner R, Wei Y, Meighen E, Derewenda Z. Biochemistry. 1994;33:9382–9388. doi: 10.1021/bi00198a003. [DOI] [PubMed] [Google Scholar]
- 13.Benning M M, Wesenberg G, Liu R L, Taylor K L, Dunaway-Mariano D, Holden H M. J Biol Chem. 1998;273:33572–33579. doi: 10.1074/jbc.273.50.33572. [DOI] [PubMed] [Google Scholar]
- 14.Schriner J E, Yi W, Hofmann S L. Genomics. 1996;34:317–322. doi: 10.1006/geno.1996.0292. [DOI] [PubMed] [Google Scholar]
- 15.Bellizzi J J, III, Widom J, Kemp C W, Clardy J. Structure (London) 1999;7:R263–R267. doi: 10.1016/s0969-2126(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 16.Szebenyi D M E, Arvai A, Ealick S, Laluppa J M, Nielsen C. J Synchrotron Radiation. 1997;4:128–135. doi: 10.1107/S0909049597000204. [DOI] [PubMed] [Google Scholar]
- 17.Leslie A G W. Mosflm User Guide, Mosflm Version 5.30. Cambridge, U.K.: MRC Lab. Mol. Biol.; 1994. [Google Scholar]
- 18.Collaborative Computational Project 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 19.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 21.Abrahams J, Leslie A. Acta Crystallogr D. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 22.Jones T, Kjeldgaard M. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Andersson S, Davids D N, Dahlback H, Jornvall H, Russell D W. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 26.Soyombo A A, Hofmann S L. J Biol Chem. 1997;272:27456–27463. doi: 10.1074/jbc.272.43.27456. [DOI] [PubMed] [Google Scholar]
- 27.Ollis D L, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken S M, Harel M, Remington S J, Silman I, Schrag J, et al. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 28.Esnouf R M. J Mol Graph Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 29.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 30.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 31.Nicholls A, Sharp K, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 32.Das A K, Becerra C H R, Yi W, Lu J-Y, Siakotos A N, Wisniewski K N, Hofmann S L. J Clin Invest. 1998;102:361–370. doi: 10.1172/JCI3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derewenda Z S, Sharp A M. Trends Biochem Sci. 1993;18:20–25. doi: 10.1016/0968-0004(93)90082-x. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Szittner R, Derewenda Z S, Meighen E A. Biochemistry. 1996;35:9967–9973. doi: 10.1021/bi9605292. [DOI] [PubMed] [Google Scholar]
- 35.Waliany S, Das A K, Gaben A, Wisniewski K E, Hofmann S L. Hum Mutat. 2000;15:206–207. doi: 10.1002/(SICI)1098-1004(200002)15:2<206::AID-HUMU14>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 36.Hofmann S L, Lee L A, Lu J-Y, Verkruyse L A. Neuropediatrics. 1997;28:27–30. doi: 10.1055/s-2007-973661. [DOI] [PubMed] [Google Scholar]
- 37.Derewenda U, Swenson L, Green R, Wei Y, Dodson G G, Yamaguchi S, Haas M J, Derewenda Z S. Nat Struct Biol. 1994;1:36–46. doi: 10.1038/nsb0194-36. [DOI] [PubMed] [Google Scholar]
- 38.Vines D J, Warburton M J. FEBS Lett. 1999;443:131–135. doi: 10.1016/s0014-5793(98)01683-4. [DOI] [PubMed] [Google Scholar]
- 39.Lerner T J, Boustany R N, Anderson J W, D'Arigo K D, Schlumpf K, Buckler A J, Gusella J F, Haines J L, Kremmidiotis G, Lensink I L, et al. Cell. 1995;82:949–957. [Google Scholar]
- 40.Jarvela I, Sainio M, Rantamaki T, Olkkonen V M, Carpen O, Peltonen L, Jalanko A. Hum Mol Genet. 1998;7:85–90. doi: 10.1093/hmg/7.1.85. [DOI] [PubMed] [Google Scholar]
- 41.Kida E, Kaczmarski W, Golabek A A, Kaczmarski A, Michalewski M, Wisniewski K E. Mol Genet Metab. 1999;66:265–271. doi: 10.1006/mgme.1999.2837. [DOI] [PubMed] [Google Scholar]
- 42.Mitchison H M, Hofmann S L, Becerra C H R, Munroe P B, Lake B D, Crow Y J, Stephenson J B P, Williams R E, Hofman I L, Taschner P E M, et al. Hum Mol Genet. 1998;7:291–297. doi: 10.1093/hmg/7.2.291. [DOI] [PubMed] [Google Scholar]