Abstract
Temporal control of p27Kip1 (p27) degradation imposes periodicity in its activity during cell cycle progression and its accumulation during cell cycle exit. Degradation of p27 is initiated by phosphorylation of p27 at Thr-187, which marks the protein for ubiquitination by SCFSkp2 and subsequent proteolysis by the 26S proteasome. Here we show that the p27 ubiquitination activity in cell extracts depends on the presence of the ubiquitin-like protein Nedd8 and enzymes that catalyze Nedd8 conjugation to proteins. Moreover, we show that reconstitution of the p27 ubiquitination activity of recombinant SCFSkp2 also requires Nedd8 conjugation pathway components. Inactivation of the Nedd8 conjugation pathway by a dominant negative mutant of the Nedd8-conjugating enzyme Nce1/Ubc12 blocks the ubiquitination and degradation of p27 in cell extracts. Consistent with a role in cell-cycle progression, Nedd8 is expressed in proliferating cells and is itself down-regulated upon cellular differentiation. These results suggest that the Nedd8 conjugation pathway may regulate the turnover of p27Kip1, independently of p27 phosphorylation, and further establishes the identity of protein components involved in p27 ubiquitination. Finally, these findings provide a direct demonstration of a function for Nedd8 in a biological process.
The cyclin-dependent kinase (CDK) inhibitor p27Kip1 (p27) functions as a negative regulator of CDKs and inactivates cyclin E/CDK2 upon mitogen starvation (1–4). The cellular activity of p27 correlates with periodic fluctuations in protein level during cell cycle progression, being highest in the G1 phase and declining rapidly as cells enter S phase (5). Increases in p27 activity in contact-inhibited and serum-starved cells also are accompanied by protein accumulation (6, 7). Changes in p27 levels primarily are caused by posttranslational events, with accumulation of p27 in G0 cells largely the result of an increase in protein stability whereas reduced levels in S phase cells are attributed in part to an increase in protein turnover (6, 8). Proper control of p27 levels is important for cell cycle progression as ectopic expression leads to cell arrest at the G1 phase (9). In addition, heterozygous p27(+/−) cells show increased susceptibility to tumorigenesis (10), and a multitude of clinical studies have correlated tumor aggressiveness and poor survival prognosis with reduced p27 protein levels or half-life (11, 12).
The degradation of p27 is mediated by ubiquitin-dependent proteolysis (13, 14) initiated by phosphorylation of Thr-187 in p27 (9, 14, 15). Phosphorylated p27 interacts with Skp2 (16–18), a member of the F-box family of proteins that generally associate with Cul1, Skp1, and Rbx1 to form multiprotein complexes known as SCFs (19). SCF complexes containing the yeast F-box proteins Cdc4 or Grr1, or the human F-box protein βTrCP, are known to function as ubiquitin-protein ligases (20–23) and are active for in vitro ubiquitination of specific phosphorylated substrate proteins. SCFSkp2 interacts specifically with p27 phosphorylated on Thr-187 (16–18); however, demonstration of p27 ubiquitination activity using complexes reconstituted from recombinant systems has met with both positive (18) and negative results (16). Here we show that p27 ubiquitination in HeLa cell extract requires the ubiquitin-like protein Nedd8 and enzymes that catalyze Nedd8 conjugation to proteins. Furthermore, we show that reconstitution of p27 ubiquitination activity of a recombinant SCFSkp2 depends on protein components in the Nedd8 conjugation pathway.
Materials and Methods
Plasmids.
The p27 cDNA was inserted into pET-21a (Novagen) between NheI and XhoI sites encoding p27 fused to Met-Ala-Ser at the N terminus and Leu-Glu-(His)6 at the C terminus. pGEX4T2/p27 was obtained by cloning the p27 cDNA between the BamHI and EcoRI sites of pGEX4T2 (Amersham Pharmacia). Mutations were generated by using the QuickChange Site-Directed Mutagenesis Kit (Stratagene).
Proteins.
Ubiquitin was purchased from Sigma. Ubiquitin aldehyde and Nedd8 aldehyde were prepared as described (24). Human Cdc34 was expressed in Escherichia coli and purified over Ni-nitrilotriacetic acid (NTA) resin (Qiagen, Chatsworth, CA), followed by size exclusion chromatography. Preparation of recombinant Nedd8, Nedd8-conjugating enzyme Nce1/Ubc12 wild type, and C111S mutant, ubiquitin-activating enzyme E1, and isolation of Nedd8-activating enzyme Nae1 from HeLa cells were as described (39). Cyclin E/CDK2 complex was expressed in Sf9 cells coinfected with recombinant baculoviruses encoding His6-tagged cyclin E and CDK2 and was purified as described (26). 35S-methionine-labeled His6-tagged p27 (wild type and T187A mutant) were prepared by coupled in vitro transcription/translation using pET-21a/p27 in a rabbit reticulocyte lysate system (TNT, Promega). Glutathione S-transferase (GST)-fused p27 (wild type, F62/64A, and T187A mutants) were expressed in E. coli by using pGEX4T2/p27 and purified on glutathione Sepharose (Amersham Pharmacia).
SCFSkp2 and Skp1/Cul1 complexes were expressed in Sf9 cells coinfected with recombinant baculoviruses encoding His6-tagged Cul1, His6-tagged Skp1, and Skp2 (for SCFSkp2), or His6-tagged Cul1 and His6-tagged Skp1 (Skp1/Cul1) and purified by using Ni-NTA resin according to ref. 16. Antibodies against Skp1 and Cul1 were purchased from Neomarker (Fremont, CA); anti-Skp2 was purchased from Zymed.
Preparation of S100 Extract, Fraction I (FI), and Fraction II (FII).
HeLa cell cytosolic extract (S100) was purchased from Cellex (Minneapolis, MN). Extract preparation included 80% ammonium sulfate precipitation and dialysis against 50 mM Tris⋅HCl (pH 7.5), 0.5 mM DTT, 0.5 mM EDTA, 0.5 mM EGTA, and 20% glycerol.
S100 extract (400 mg) was loaded onto a 20-ml Q Sepharose Fast Flow column (Amersham Pharmacia) equilibrated in buffer A (50 mM Hepes/Na, pH 7.6/1 mM DTT). The unbound proteins were pooled and precipitated with 70% ammonium sulfate (FI). Bound proteins were eluted with 0.5 M NaCl in buffer A and precipitated with 60% ammonium sulfate (FII). Both FI and FII were redissolved in buffer A, dialyzed against the same buffer, and stored at −80°C.
[32P]-Phosphorylation of GST-p27 with Cyclin E/CDK2.
GST-p27 F62/64A (2.1 μg) was phosphorylated for 30 min at 30°C in a 100-μl reaction containing 50 mM Hepes/Na (pH 7.6), 6 mM MgCl2, 1 μM okadaic acid (GIBCO/BRL), 10 μM [γ-32P]ATP (150 Ci/mmol), and 0.4 μg of cyclin E/CDK2. [32P]GST-p27 was repurified by using glutathione Sepharose (Amersham Pharmacia).
In Vitro Ubiquitination Assay.
Reaction mixtures (20 μl) were typically incubated for 1 h at 30°C and contained 10 mM Hepes/Na (pH 7.6), 6 mM MgCl2, 2 mM ATP, 1 μM okadaic acid, 30 μM ubiquitin, 2 μM ubiquitin aldehyde, 4 μM proteasome inhibitor MG273 (LeukoSite), 0.5 μl of in vitro-translated [35S]His6-p27, 1 μl of in vitro-phosphorylated [32P]GST-p27, or 50 ng of unlabeled GST-p27. Reactions containing unphosphorylated GST-p27 also contained 20 ng of cyclin E/CDK2. S100 extract (80 μg), FI (20 μg), or FII (30 μg) were included as specified in the figure legends. [35S]His6-p27 and its polyubiquitinated forms were purified from the reaction mixtures with Ni-NTA resin before SDS/PAGE. Reactions using radiolabeled p27 were analyzed by using a Storm PhosphorImager (Applied Biosystems). Nonradioactive GST-p27 was analyzed by Western blotting using a monoclonal anti-p27 antibody (Transduction Laboratories, Lexington, KY).
Identification of Nedd8.
FI (0.5 ml, 5 mg protein) was adjusted to pH 9.3 and loaded onto a MonoP HR 5/20 column (Amersham Pharmacia) equilibrated in 75 mM Tris/acetate, pH 9.3. Proteins were eluted with 10% Polybuffer 96 adjusted to pH 6.0 with acetic acid, and fractions were assayed for ability to reconstitute the polyubiquitination of [35S]p27 in the presence of FII. The activity eluted in fractions with pH 7.5–7.7. These fractions were combined, concentrated, and chromatographed on a Superdex Peptide HR 10/30 column (Amersham Pharmacia) equilibrated in 20 mM Hepes/Na (pH 7.6), 0.5 mM DTT. Fractions were collected and assayed for activity. Active fractions (9.6- to 10.8-ml elution volume) were analyzed by SDS/PAGE, and a single protein species with an apparent molecular mass of 8–9 kDa was enriched in the active fractions. MS and peptide sequence analysis of this protein were carried out at the fee-for-service Microsequencing Facility at Emory University (Atlanta, Georgia).
Results
Nedd8 Is Required for p27 Ubiquitination.
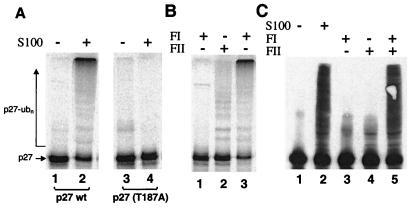
To identify proteins that participate in p27 ubiquitination, we reconstituted ubiquitination activity using fractionated cell extracts and recombinant proteins. Ubiquitination of in vitro-translated 35S-labeled p27 was detected by the appearance of high molecular weight conjugates of the radiolabeled p27 in SDS gels. We used asynchronous HeLa cell extract as the starting material for protein fractionation after determining that an S100 extract from such cells could fully support the ubiquitination of p27 in vitro (Fig. 1A, lane 2). The ubiquitination and degradation of p27 in vivo is known to be initiated by phosphorylation of amino acid Thr-187 in this protein, and substitution of Thr-187 to alanine is known to stabilize p27 in cells but does not alter its ability to inhibit CDKs (9, 15). This phosphorylation requirement was retained in this in vitro reaction system because a p27-T187A mutant was not subject to ubiquitination in the S100 extract (Fig. 1A, lane 4).
Figure 1.
In vitro ubiquitination of p27Kip1. (A) HeLa S100 contains a ubiquitin ligase activity specific for p27 phosphorylated on Thr-187. Reaction mixtures contained S100 extract as indicated and [35S]His6-p27 wild type (lanes 1 and 2) or mutant (T187A) (lanes 3 and 4). After incubation for 1 h at 30°C, [35S]His6-p27 and its polyubiquitinated forms were resolved on a 12% Tris-glycine SDS-gel, and analyzed by phosphorimaging. (B) Both FI and FII are required for robust ubiquination of in vitro-translated p27. [35S]His6-p27 was incubated with FI and/or FII as indicated, and reactions were analyzed as in A. Note that FII alone supported a basal level of ubiquitination (lane 2), which was likely caused by reticulocyte lysate in the reaction. (C) Ubiquitination of bacterially expressed p27 is FI and FII dependent. Reactions contained S100 extract, FI, and/or FII as indicated. GST-p27 and its polyubiquitinated forms were resolved on an 8% Tricine SDS-gel and immunoblotted by using anti-p27 antibodies. Note that robust p27 conjugate formation was detected only in reactions containing S100 or both FI and FII (lanes 2 and 5).
Fractionation of HeLa S100 proteins was carried out to initially generate two protein pools based on retention on an anion exchange resin. FI and FII were obtained by pooling proteins that were either not retained by Q-Sepharose (FI) or that were eluted by 0.5 M NaCl (FII), respectively. These two fractions then were tested individually or in combination for p27 ubiquitination activity. Ubiquitination of proteins requires the activation of ubiquitin by the ubiquitin-activating enzyme (E1), and the concerted action of a ubiquitin-conjugating enzyme (E2) and a ubiquitin-protein ligase (E3) (25). To restrict our efforts to the identification of E2, E3, and possibly other novel components, fractionated HeLa proteins were assayed for p27 ubiquitination activity in reactions supplemented with purified E1, ubiquitin, and cyclin E/CDK2, which provides the kinase activity for phosphorylation of Thr-187 in p27 (26). Reconstitution of robust ubiquitination activity required both FI and FII together (Fig. 1B, lane 3), although a minor activity could be detected by using only FII, but not FI alone (Fig. 1B, lanes 1 and 2). Because the small quantity of rabbit reticulocyte lysate in the p27 translation mix could contribute to the activity observed with FII alone, we repeated the assays with bacterially expressed GST-p27 as substrate (Fig. 1C). Ubiquitination of this substrate also required both FI and FII proteins (Fig. 1C, lane 5). Importantly, FII alone was essentially devoid of activity in these reactions as evidenced by the lack of detectable high molecular weight conjugates (Fig. 1C, lane 4), suggesting that the activity observed with the p27 translation mix was indeed likely caused by reticulocyte lysate in the reaction.
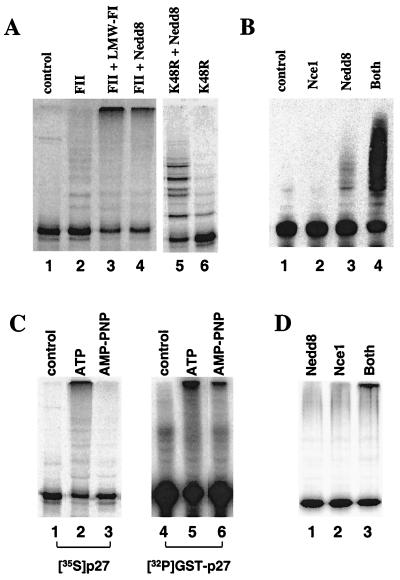
We next subjected FI and FII proteins to gel filtration chromatography on Superdex G-75 and tested the resulting fractions for the ability to replace either FI or FII in ubiquitination reactions. This analysis revealed that FI could be replaced by FI proteins of apparent molecular mass between 8 and 14 kDa when assayed by using the p27 translation mix (Fig. 2A, lane 3), but not with the GST-p27 fusion protein (data not shown). These results were consistent with a model in which FI contained more than one essential component of which only one was limiting when reticulocyte lysate was included in the reaction. We further purified this FI factor by sequential chromatofocusing and size exclusion chromatography and assigned the activity to a single protein that migrated on SDS gels with an apparent molecular mass of 9 kDa (data not shown). After excision of this protein from an SDS gel, peptide fragments were obtained by digestion with trypsin and purification by HPLC. One peptide with a molecular mass of 1,686 Da as determined by MS was analyzed further by amino acid sequencing, yielding the sequence EIEIDIEPTDKVER, which is identical to residues 12–25 of the 76-residue protein Nedd8 (GenBank accession no. Q15843). We next expressed and purified recombinant human Nedd8 from bacteria for testing in ubiquitination reactions. Indeed, the recombinant protein when added to FII restored the ubiquitination of in vitro-translated p27 (Fig. 2A, lane 4). To confirm that the high molecular weight p27 conjugates formed in these assays were in fact p27-ubiquitin conjugates, we carried out similar reactions replacing ubiquitin with the ubiquitin mutant Ub-K48R, which is defective in polyubiquitin chain assembly (27). With Ub-K48R, p27 conjugate formation also required Nedd8 but only discrete low molecular weight conjugates were formed (Fig. 2A, lanes 5 and 6), consistent with the expectation that polyubiquitin chain formation was disrupted by the mutation in Ub-K48R.
Figure 2.
Nedd8 is required for p27 ubiquitination. (A) Ubiquitination of p27 requires a FI component. In vitro-translated [35S]His6-p27 was incubated in the absence (lane 1) or presence of FII (lanes 2–6). FII containing reactions were further supplemented by the addition of buffer alone (lane 2), a low molecular weight gel filtration fraction of FI (lane 3), or 0.29 μM Nedd8 (lane 4). Substitution of ubiquitin with Ub-K48R results in the formation of discrete low molecular weight p27-ubiquitin conjugates in the presence (lane 5) but not the absence of recombinant Nedd8 (lane 6). (B) Ubiquitination of p27 by FII requires both Nedd8 and the Nedd8-conjugating enzyme, Nce1. GST-p27 was incubated with FII alone (lane 1) or in the presence of 1 μM Nce1 and/or 0.29 μM recombinant Nedd8 as indicated (lanes 2–4). (C) Prior phosphorylation is required for p27 ubiquitination. Reactions were performed by using S100 extract as a source of ubiquitinating enzymes, and either in vitro-translated [35S]His6-p27 or bacterially expressed GST-p27 as substrates. Ubquitination of [35S]His6-p27 (lanes 1–3) was carried out in the absence (lane 1) or presence of 2 mM ATP (lane 2) or 2 mM AMP-PNP (lane 3). In lanes 4–6, GST-p27 (F62/64A) was prephosphorylated with cyclin E/CDK2 using [γ-32P]ATP and repurified. Reactions containing [32P]-GST p27 were conducted in the absence (lane 4) or presence of 2 mM ATP (lane 5) or 2 mM AMP-PNP (lane 6). (D) Ubiquitination of prephosphorylated [32P]-GST-p27 by FII requires both Nedd8 and Nce1 (lanes 1–3).
Nedd8 Activity in p27 Ubiquitination Requires the Nedd8-Conjugating Enzyme Nce1.
Nedd8 and ubiquitin belong to a family of proteins whose C termini can be linked to other cellular proteins (28). Nedd8 conjugation requires distinct activating and conjugating enzymes with similar functions as their counterparts in the ubiquitin pathway (28, 29). Independent experiments indicated that Nedd8-activating enzyme, Nae1, is present in FII, whereas the Nedd8-conjugating enzyme, Nce1, is present in FI (T.B.G. and V.C., unpublished results). Nce1 is identical to Ubc12, a protein shown to function as a Nedd8-conjugating enzyme in rabbit reticulocyte lysate (30). The presence of this activity in reticulocyte lysate could explain why Nedd8 alone was sufficient to replace FI in assays with in vitro-translated p27, but not with the bacterially expressed GST- p27 protein. To investigate the possibility that Nce1 is required for p27 ubiquitination, we tested purified Nce1 protein that was expressed in bacteria. Ubiquitination of GST-p27 (Fig. 2B) was tested with FII (lane 1), which was supplemented with either Nce1 (lane 2), Nedd8 (lane 3), or both (lane 4). While a small amount of low molecular weight p27 conjugates were detected with added Nedd8, the inclusion of both Nedd8 and Nce1 led to a robust reaction as well as the formation of high molecular weight p27 conjugates. When ubiquitin was omitted from the reaction mixture, p27 conjugate formation was not detected under any of these conditions (data not shown), indicating that p27 conjugates are formed exclusively with ubiquitin and not with Nedd8.
Nedd8 Is Required in the Ubiquitination Step.
Ubiquitin-mediated degradation is targeted by phosphorylation at residue Thr-187 of p27, therefore Nedd8 could be required either at phosphorylation or at a subsequent step during ubiquitination. To distinguish these possibilities, we assayed the ubiquitination step using prephosphorylated GST-p27 in reaction mixtures in which ATP was replaced by adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP) and using S100 as a source of ubiquitinating enzymes. Because additional p27 phosphorylation could not occur in the absence of ATP, the reaction in the presence of AMP-PNP is limited to the ubiquitination of the prephosphorylated substrate protein. As expected, in ATP-containing reactions, no requirement for prephosphorylation could be demonstrated (Fig. 2C, lanes 2 and 5). However, when AMP-PNP was used instead of ATP, p27 ubiquitination depended on prephosphorylation of p27 (Fig. 2C, compare lanes 3 and 6). Importantly, FII could support ubiquitination of prephosphorylated p27 only in the presence of both Nedd8 and Nce1 (Fig. 2D, lane 3), indicating that Nedd8 and Nce1 are required for the ubiquitination step.
Reconstitution of Recombinant SCFSkp2 Ubiquitination Activity Requires the Nedd8 Pathway.
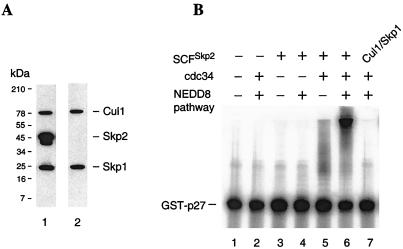
Although our studies so far have not addressed the identity of FII proteins, one essential component is expected to be the Nedd8-activating enzyme, Nae1. However, FII also contains additional component(s) because neither the E2 nor the E3 enzymes for p27 ubiquitination are FI proteins, and purified Nae1 was insufficient to replace FII in p27 ubiquitination reactions (data not shown). Two recent studies have suggested an essential role for the SCFSkp2 ubiquitin-protein ligase complex in ubiquitin-mediated degradation of p27 (16, 17), and we have determined by immunoblotting with specific antibodies that the Cul1, Skp1, and Skp2 components of this complex are found exclusively in FII (data not shown). We thus tested directly whether FII could be replaced in our assay by the addition of Nae1, a recombinant form of the SCFSkp2 complex, and the ubiquitin-conjugating enzyme Cdc34 (19). We coexpressed the subunits of SCFSkp2 in Sf9 cells by using recombinant baculoviruses encoding of His6-tagged Cul1, His6-tagged Skp1, and Skp2, and purified the complex by Ni-affinity chromatography. The copurification of all three proteins was confirmed by immunoblotting (Fig. 3A, lane 1). Recent studies indicated that SCF complexes also contain the ring finger protein Rbx1/Roc1 (32–34). Although not specifically tested here, the presence of this protein in a similar preparation was confirmed by immunoblotting with Roc1-specific antibodies (M. Pagano, personal communication). Apparently the recombinant Cul1/Skp1/Skp2 complex efficiently recruited the host Rbx1/Roc1 subunit. We also obtained a complex derived by coexpressing only Cul1 and Skp1 and confirmed the absence of Skp2 by immunoblotting (Fig. 3A, lane 2). These complexes then were analyzed for p27 ubiquitination activity. The SCFSkp2 complex, but not the complex lacking Skp2, was clearly active when assayed in the presence of both Cdc34 and protein components of the Nedd8 conjugation pathway (Fig. 3B, lanes 6 and 7). Similar to results with FII shown in Fig. 2B, p27 conjugates formation with the SCFSkp2 complex also entirely depends on the presence of added ubiquitin (data not shown). In the absence of the Nedd8 conjugation components, the SCFSkp2 complex was largely devoid of activity although a low level of p27 conjugates could be detected (Fig. 3B, lane 5). This latter result is consistent with previous studies that used similar preparation of SCFSkp2 complex that was shown to interact specifically with phosphorylated p27, but was either inactive or partially active for ubiquitination activity (16, 18). In those studies, the authors speculated that components essential for ubiquitination activity could be missing from the assembled, recombinant SCFSkp2. Our results here suggest that components of the Nedd8 conjugation pathway, including Nedd8, Nce1, and Nae1, are required for SCFSkp2-mediated ubiquitination of p27.
Figure 3.
Reconstitution of p27 ubiquitination activity. (A) SCFSkp2 and Cul1/Skp1 complexes were prepared from Sf9 cells coexpressing His6-tagged Cul1, His6-tagged Skp1, and Skp2 (lane 1), or His6-tagged Cul1 and His6-tagged Skp1. Proteins were purified with Ni-NTA resin and analyzed by SDS/PAGE and immunostaining with antibodies against Cul1, Skp1, and Skp2. (B) Ubiquitination reactions containing [32P]-GST-p27, 30 μM ubiquitin, 100 nM E1 (ubiquitin activating enzyme), 2 μM ubiquitin aldehyde, 4 μM MG273, 1 μM okadaic acid, and 2 mM ATP were carried out by using in the absence (lanes 1 and 2) or presence of SCFSkp2 (lanes 3–6) or Cul1/Skp1 (lane 7). Reaction mixtures were further supplemented with Nedd8 pathway components (0.29 μM Nedd8, 1 μM Nce1, and 100 nM Nae1; lanes 2, 4, 6, and 7) and 1 μM Cdc34 (lanes 2 and 5–7).
A Catalytic-Site Mutant of Nce1 Exerts a Dominant Negative Effect.
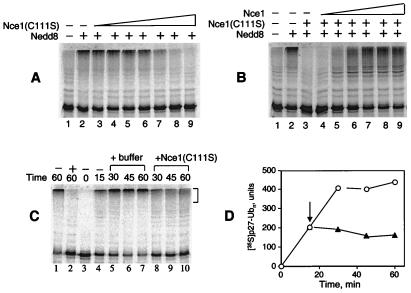
The requirement of Nedd8 conjugation in p27 ubiquitination was demonstrated further by the use of a catalytic site mutant of Nce1. Substitution of the active site cysteine in ubiquitin-conjugating enzymes has been used to generate dominant negative mutants (35, 36), and we engineered a similar mutation by replacing Cys-111 in Nce1 with a serine. The mutant was expressed in bacteria, purified, and tested for the ability to block p27 ubiquitination in HeLa S100 extracts. Addition of the Nce1(C111S) mutant blocked p27 ubiquitination in a dose-dependent manner (Fig. 4A). The inhibitory effect of Nce1(C111S) could be reversed in these reactions by inclusion of wild-type Nce1 (Fig. 4B), suggesting that Nce1 (C111S) inhibition was not caused by an indirect effect.
Figure 4.
The Nce1 C111S mutant exerts a dominant negative effect on p27 ubiquitination. (A) Ubiquitination of in vitro-translated [35S]His6-p27 was carried out with FII, 0.29 μM Nedd8 (lanes 2–9), and Nce1 mutant C111S in increasing concentrations: 0.08, 0.2, 0.3, 0.5, 1, 2, and 4 μM (lanes 3–9). (B) Wild-type Nce1 reverses inhibition by Nce1 C111S. Ubiquitination of in vitro-translated [35S]His6-p27 was carried out by using FII, 0.29 μM Nedd8 (lanes 2–9), 4 μM Nce1 C111S (lanes 3–9), and increasing wild-type Nce1: 0.02, 0.1, 0.2, 0.5, 1, and 2 μM (lanes 4–9, respectively). (C) Continuous Nce1 activity is required to sustain the p27 ubiquitination reaction. Ubiquitination of in vitro-translated [35S]His6-p27 was carried out by using S100 extract. Two reactions were incubated in the absence (lane 1) or presence (lane 2) of 4 μM Nce1 C111S for 60 min. The reaction in lane 3 was immediately mixed with Ni-NTA beads and kept on ice until completion of the experiment. The remaining reactions were incubated at 30°C for 15 min and either terminated by the addition of Ni-NTA and chilling on ice (lane 4), or mixed with Nce1 C111S (final concentration 4 μM, lanes 8–10), or mock buffer (lanes 5–7). Reactions were further incubated 30°C for an additional 15, 30, or 45 min before termination. (D) Polyubiquitinated [35S]p27 reaction products (shown with brackets in C) were quantified by phosphorimage analysis. The arrow indicates the addition of Nce1 C111S (▴) or mock buffer (○).
The Nce1(C111S) mutant affects on p27 ubiquitination suggested that continuous Nedd8 conjugation activity is required to sustain the p27 ubiquitination reaction. Results shown in Fig. 4C indicate that p27 ubiquitination activity in HeLa S100 was rapidly inactivated by the addition of Nce1(C111S), either before or during the reaction (Fig. 4 C and D). Consistent with these results, preincubation of a mixture containing FI and FII in the presence of ATP did not affect the ability of Nce1(C111S) to inhibit p27 ubiquitination (data not shown). To address the possibility that Nedd8 could itself form conjugates directly with p27, we performed reactions using FII, Nce1/Ubc12, ATP, and 125I-labeled Nedd8 and did not detect the formation of Nedd8-p27 conjugates (data not shown). Under the same conditions, modification by Nedd8 was observed for >70% of an in vitro-translated Cul2, a known substrate of Nedd8 conjugation (data not shown). Thus, the requirement for the continuous presence of Nedd8 conjugation activity to sustain p27 ubiquitination cannot be explained by the simple mechanism involving p27 modification by Nedd8.
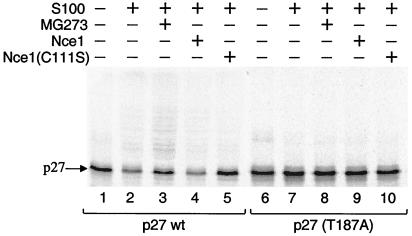
We next carried out reactions to monitor p27 degradation in HeLa S100 extracts under conditions similar to those for ubiquitination, but excluding proteasome inhibitor and ubiquitin aldehyde, an inhibitor of isopeptidase activities that disassemble ubiquitin–protein conjugates. Degradation of p27 under these conditions required the presence of Thr-187 (Fig. 5), consistent with results from previous reports (9, 26). Inactivation of 26S proteasome by the addition of the inhibitor MG273 led to stabilization of p27 and accumulation of ubiquitinated p27 (Fig. 5, lane 2), consistent with ubiquitination being a prerequisite for its degradation. p27 also was stabilized by the addition of the Nce1(C111S) mutant (Fig. 5, lane 5), consistent with our findings that Nedd8 conjugation is required for p27 ubiquitination. Taken together, our results raise the interesting possibility that the turnover of cellular p27 may be sensitive to changes in the activity of the Nedd8 conjugation pathway, in addition to p27 phosphorylation status.
Figure 5.
Ubiquitin-dependent degradation of p27 requires Thr-187, Nedd8-conjugating activity, and proteasome activity. Ubiquitination reactions were performed by using in vitro-translated [35S]His6-p27 (lanes 1–5) or mutant (T187A) (lanes 6–10) in the presence or absence of S100 extract, proteasome inhibitor MG273, wild-type Nce1, or Nce1 C111S as indicated.
Discussion
Phosphorylation of p27 on Thr-187 targets this CDK inhibitor for degradation via the ubiquitin proteasome pathway. Recent reports have established a role for cyclin E/CDK2 in p27 phosphorylation (14, 15, 26) and have identified SCFSkp2 as an E3 ligase that specifically recognizes this phosphorylated substrate (16–18). SCFSkp2 is composed of multiple components including Cul1, Skp1, and the F-box protein, Skp2. Here we established an in vitro reaction system that retained the Thr-187 phosphorylation requirement and enabled us to identify the essential participation of Nedd8 and enzymes of the Nedd8 conjugation pathway in p27 ubiquitination by several criteria. First, we identified Nedd8 as a protein required for p27 ubiquitination in asynchronous human cell extracts, and we confirmed this result by using bacterially expressed recombinant Nedd8. Secondly, we showed that p27 ubiquitination activity in human cell extract also requires Nce1/Ubc12, a known Nedd8-conjugating enzyme. Moreover, we were able to faithfully reconstitute p27 ubiquitination activity by using recombinant SCFSkp2 only when the Nedd8 pathway components Nedd8, Nce1, and Nae1 were included in reactions. Finally, we showed that an active site mutant of Nce1 exerts a dominant negative effect on p27 ubiquitination and degradation.
We have established that Nedd8 modification is not required for p27 phosphorylation, and the absence of detectable Nedd8-p27 conjugates suggests that Nedd8 modifies a protein other than p27 itself. Further insights into the role of Nedd8 in p27 ubiquitination will require definitive identification of the Nedd8 acceptor protein. However, given that Nedd8 is known to modify Cdc53 and cullin family members in ubiquitin-protein ligases (29–31, 37–39), one obvious candidate for the Nedd8 acceptor protein is the Cul1 component of SCFSkp2. Because p27 ubiquitination activity is rapidly inactivated by the Nce1 dominant-negative mutant, modification of the ligase by Nedd8 is predicted to be a dynamic process with profound affects on the activity of the enzyme. Conjugates formed with ubiquitin and SUMO1 are subject to disassembly by isopeptidases (40). Although an analogous isopeptidase has not been described for Nedd8-modified proteins, the existence of this activity could result in a dynamic process requiring the continuous presence of Nedd8 conjugation activity. A prediction of this model is that once the Nedd8-conjugated form of SCFSkp2 is generated, SCFSkp2-mediated ubiquitination of p27 in the absence of the hypothetical Nedd8 isopeptidase should no longer require the continuous presence of Nedd8 pathway components. To date, our attempts to test this idea by using recombinant components have not yielded an SCFSkp2 complex that is active in the absence of Nedd8-conjugating activity (data not shown). As an alternative test of this hypothesis, we generated Nedd8 aldehyde (analogous to ubiquitin aldehyde) as an inhibitor of Nedd8 isopeptidase activity. Addition of this reagent to ubiquitation reactions also failed to relieve the requirement for Nedd8 pathway components (data not shown). Thus, although the mechanistic function of Nedd8 in p27 ubiquitination remains to be defined, our results nonetheless suggest a biological function for Nedd8 in cullin containing ubiquitin ligases.
Our findings imply that p27 levels may be regulated by the activity of the Nedd8 conjugation pathway, coordinately with the status of Thr-187 phosphorylation. Nedd8 originally was identified as a protein whose expression is developmentally down-regulated (41), and Nedd8 protein levels are reduced in differentiated versus proliferating cells (42). Whether the level of Nedd8 plays a regulatory role in its function is an issue under investigation. Indeed, the enzymes that function in Nedd8 conjugation may be subject to similar regulation. Finally, Nedd8 conjugation to protein is likely subject to regulation. The observation that Nedd8 is attached to only a small percentage of cellular Cdc53/Cul1 and Cul2 suggests that protein modification by Nedd8 is a regulated process, albeit by yet to be defined mechanism(s).
Acknowledgments
We thank M. Pagano for baculoviruses that express cyclin E, CDK2, Cul1, Skp1, and Skp2 and for communication of results before publication. We thank L. Dang for Nedd8 aldehyde and M. Read, V. Palombella, and J. Adams for carefully reading the manuscript. This work was supported in part by National Institutes of Health Grant GM53136.
Abbreviations
- CDK
cyclin-dependent kinase
- SCF
skp1, cul1, F-box protein complex
- NTA
nitrilotriacetic acid
- GST
glutathione S-transferase
- FI
fraction I
- FII
fraction II
- AMP-PNP
5′-[β,γ-imido]triphosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090465597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090465597
References
- 1.Sherr C J, Roberts J M. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 2.Elledge S J, Harper J W. Curr Opin Cell Biol. 1994;6:847–852. doi: 10.1016/0955-0674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 3.Reynisdottir I, Polyak K, Iavarone A, Massague J. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 4.Clurman B E, Porter P. Proc Natl Acad Sci USA. 1998;95:15158–15160. doi: 10.1073/pnas.95.26.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng M, Olivier P, Diehl J A, Fero M, Roussel M F, Roberts J M, Sherr C J. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hengst L, Reed S I. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- 7.Polyak K, Kato J Y, Solomon M J, Sherr C J, Massague J, Roberts J M, Koff A. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 8.Pagano M. FASEB J. 1997;11:1067–1075. doi: 10.1096/fasebj.11.13.9367342. [DOI] [PubMed] [Google Scholar]
- 9.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 10.Fero M L, Randel E, Gurley K E, Roberts J M, Kemp C J. Nature (London) 1998;396:177–180. doi: 10.1038/24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsihlias J, Kapusta L, Slingerland J. Annu Rev Med. 1999;50:401–423. doi: 10.1146/annurev.med.50.1.401. [DOI] [PubMed] [Google Scholar]
- 12.Loda M, Cukor B, Tam S W, Lavin P, Fiorentino M, Draetta G F, Jessup J M, Pagano M. Nat Med. 1997;3:231–234. doi: 10.1038/nm0297-231. [DOI] [PubMed] [Google Scholar]
- 13.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Del Sal G, Chau V, Yew P R, Draetta G F, Rolfe M. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen H, Gitig D M, Koff A. Mol Cell Biol. 1999;19:1190–1201. doi: 10.1128/mcb.19.2.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlach J, Hennecke S, Amati B. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrano A C, Eytan E, Hershko A, Pagano M. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 17.Sutterlüty H, Chatelain E, Wirbelauer C, Sentfen M, Müller U, Krek W. Nat Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- 18.Tsvetkov L M, Yeh K-H, Lee S-J, Sun H, Zhang H. Curr Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- 19.Bai C P, Sen K, Hofmann L, Ma M, Goebl J W, Elledge S J. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 20.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 21.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 22.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning A M, Andersen J S, Mann M, Mercurio F, Ben-Neriah Y. Nature (London) 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 23.Latres E, Chiaur D S, Pagano M. Oncogene. 1999;18:849–854. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- 24.Hershko A, Rose I A. Proc Natl Acad Sci USA. 1987;84:1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickart C. In: Ubiquitin and Biology of the Cell. Peters J-M, Harris J R, Finley D, editors. New York: Plenum; 1998. pp. 19–63. [Google Scholar]
- 26.Montagnoli A, Fiore F, Eytan E, Carrano A C, Draetta G F, Hershko A, Pagano M. Genes Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chau V, Tobias J, Bachmair A, Marriott D, Ecker D, Gonda D, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 28.Hochstrasser M. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 29.Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A. Proc Natl Acad Sci USA. 1999;96:5510–5515. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lammer D, Mathias N, Laplaza J M, Juang W, Liu Y, Callis J, Goebl M, Estelle M. Genes Dev. 1999;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, et al. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 33.Ohta T, Michel J J, Schottelius A J, Xiong Y. Mol Cell. 1999;4:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 34.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 35.Banerjee A, Deshaies R J, Chau V. J Biol Chem. 1995;270:26209–26215. doi: 10.1074/jbc.270.44.26209. [DOI] [PubMed] [Google Scholar]
- 36.Townsley F M, Aristarkhov A, Beck S, Hershko A, Ruderman J V. Proc Natl Acad Sci USA. 1997;94:2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Oncogene. 1999;18:6829–6834. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- 38.Wada H, Yeh E T, Kamitani T. Biochem Biophys Res Commun. 1999;257:100–105. doi: 10.1006/bbrc.1999.0339. [DOI] [PubMed] [Google Scholar]
- 39.Read M A, Brownell J E, Gladysheva T B, Hottelet M, Parent L A, Coggins M B, Pierce J W, Podust V N, Luo R-S, Chau V, Palombella V J. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S J, Hochstrasser M. Nature (London) 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Yoshida Y, Noda M. Biochem Biophys Res Commun. 1993;195:393–399. doi: 10.1006/bbrc.1993.2056. [DOI] [PubMed] [Google Scholar]
- 42.Kamitani T, Kito K, Nguyen H P, Yeh E T H. J Biol Chem. 1997;272:28565–28571. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]